Clinical Medical
Reviews and Case Reports
Features of Colon Cancer with Liver Metastasis: A Case Report and Literature Review
Ruijun Bao1 and Lijiang Ma2*
1Division of Gastroenterology and Hepatology, Mount Sinai Hospital, USA
2Division of Molecular Genetics, Department of Pediatrics, Columbia University Medical Center, USA
*Corresponding author: Lijiang Ma, Division of Molecular Genetics, Department of Pediatrics, Columbia University Medical Center, New York, NY 10032, USA, Tel: 212-851-5318, Fax: 212-851-5306, E-mail: lm2689@columbia.edu
Clin Med Rev Case Rep, CMRCR-2-025, (Volume 2, Issue 4), Case Report; ISSN: 2378-3656
Received: December 17, 2014 | Accepted: March 26, 2015 | Published: April 01, 2015
Citation: Bao R, Ma L (2015) Features of Colon Cancer with Liver Metastasis: A Case Report and Literature Review. Clin Med Rev Case Rep 2:025. 10.23937/2378-3656/1410025
Copyright: © 2015 Bao R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
According to American Cancer Society, there estimated to have one million new colorectal cancer cases worldwide each year, and half of them die from clinical complications and metastasis. The dominant metastasis site for patient with colon cancer is liver. In this report, a 77-year-old male presented with occult blood in stool, weight loss and abnormal liver function tests. Previous medical history of the patient included upper GI bleeding due to duodenum ulcer and alcohol drinking for many years. Abdominal CT scan indicated obstructive sigmoid colon lesion with multiple liver metastasis. Colonoscopy revealed a nearly obstructed lesion in sigmoid colon. Palliative colostomy was performed and sigmoid mass was resected. Pathology examination demonstrated that tumor was originated from mucosa and invades through submucosa, muscular propria and pericolonic fat. Tumor cells showed moderately differentiated, high nuclear grade with calcification, necrosis and vascular invasion. The TNM staging was used and the American Joint Committee on Cancer (AJCC) tumor staging was 2a (pT3N0Mx). This case presented typical pathology features of colon cancer relate to liver metastasis. Life time alcohol consumption is one of the risk factors for colon cancer liver metastasis. Besides behavior and life style, genetics and epigenetic factors play important roles in liver metastasis in colorectal carcinoma patients. Colon cancer is an insidious disease and almost half of the cases in the United States are diagnosed at late stages. Metastasis to the liver is leading cause of death in patients with colon cancer. Epigenetic biomarkers, such as circulating factors in serum, are developed for noninvasive molecular diagnosis of colon cancer at early stage as well as colon cancer with liver metastasis. Genetics and epigenetics studies provided molecular adjuvant therapy regimens for metastasis and evaluation for prognosis.
Keywords
Colon cancer, Liver metastasis, Alcohol, Genetics, Epigenetics
Abbreviations
AJCC: American Joint Committee on Cancer, ANXA3: Annexin A3, BRAF: v-raf murine sarcoma viral oncogene homolog B, CCL8: Chemokine CC motif, Ligand 8, CLEC4D: C-type Lectin Domain Family 4. Member D, CT: Computed Tomography, DCLK1: Doublecortin-like Kinase 1, DLC1: Deleted in Liver Cancer 1, DPYS: Dihydropyrimidinase, EGFR: Epithelial Growth Factor Receptor, EMT: Epithelial�to-Mesenchymal Transition 5-Fluorouracil (5-FU), GALR2: Galanin Receptor 2, GPSM1: G-Protein Signaling Modulator 1, HE stain: Hematoxylin and Eosin Stain, IL2RB: Interleukin 2 Receptor, Beta, KRAS: Kirsten Rat Sarcoma, LMNB1: Lamin B1, MAGEA8: Melanoma Antigen Family A, 8, NEUROG1: Neurogenin1, NKX2-5: NK2 Homeobox 5, OR51B4: Olfactory Receptor, Family 51, Subfamily B, Member4, PRRG4: Proline Rich Gla (G-carboxyglutamic acid) 4 (transmembrane), RUNX3: Runt-related Transcription Factor 3, SALL1: Spalt-like Transcription Factor, SALL3: Spalt-like Transcription Factor 3, SEPT9: septin 9, SPOCK2: Sparc/osteonectin, CWCV, and kazal-like domains proteoglycan 2, SLC16A12: Solute Carrier Family 16, member 12, TMEFF2: Transmembrane Protein with EGF-like and two Follistatin-like Domains 2, TNFAIP6: Tumor Necrosis Factor, Alpha-Induced Protein 6. VNN1: Vanin 1, VEGFR: Vascular Endothelial Growth Factor Receptor, ZBTB32: Zinc Finger and BTB Domain-Containing Protein 32, ZEB2: Zinc Finger E Box-Binding Homeobox 2
Introduction
According to American Cancer Society, they estimated to have one million new colorectal cancer cases worldwide each year. Half of them die from clinical complications and metastasis. The dominant metastasis site for patient with colon cancer is liver. In this report, a case of colon cancer with liver metastasis is presented. Pathology features of tumor tissue were presented and these features are useful for surgical strategy. Patient had history of daily drinking which provided added evidence that alcohol is a risk factor for colon cancer development and liver metastasis. Patients who have history of alcohol consumption should have careful examination of liver for distant cancer dissemination. Despite progress was made in investigating colorectal tumor genesis, understanding of liver metastasis process remains unclear. Understanding the mechanisms of colon cancer liver dissemination will provide principles for the diagnosis, treatment and prevention of advanced colon cancer. Findings concerning epigenetic regulation and genetic mechanisms of colon cancer were reviewed in this report. Epigenetic and molecular biomarkers are being developed for diagnosis and therapy and are being used in clinical cancer patients.
Case Report
Patient presentation
A 77-year-old gentleman presented to the clinic with chief complains of weight loss, occult blood in stool, and abnormal liver function tests. Abdominal Computed Tomography (CT) scan revealed obstructive sigmoid colon lesion with liver metastasis (Figure 1A,1B). His previous history was insignificant except for upper gastrointestinal bleeding due to duodenum ulcer. The patient did not smoke any tobacco but has been drinking medium a day for many years.
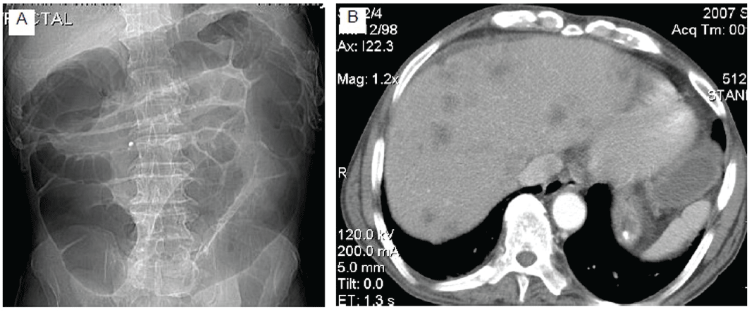
Figures 1: Abdominal CT scan demonstrating entirely dilated colon and looping in sigmoid colon (Figure 1A). CT image shows multiple low density areas in the liver (Figure 1B).
View Figures 1
Colon resection
Surgery was performed for palliative treatment. During operation, colonoscopy revealed a nearly obstructed lesion in sigmoid colon. Exploratory laparotomy demonstrated the cecum is dilated to approximately 12 centimeter with the entire transverse and descending colon also dilated to the sigmoid colon where a large mass measuring approximately 4 centimeter to be adherent to the anterior abdominal wall in the left lower quadrant. The sigmoid mass was resected and colostomy was performed.
Pathology and cancer staging
The partial colectomy specimen contains firm, thickened obstructed circumferential lesion measuring 6�5�4 centimeter with polypoid appearance in inner surface. Hematoxylin and eosin staining shows the tumor originated from mucosa (Figure 2A) and invades through submucosa (Figure 2B), muscular propria (Figure 2C) and pericolonic fat (Figure 2D). Tumor cells shows moderately differentiated, high nuclear grade (Figure 2E,2F) with calcification, necrosis (Figure 2G) and vascular invasion (Figure 2H). No carcinoma was seen in resection margins and lymph nodes. The TNM staging was used and the American Joint Committee on Cancer (AJCC) tumor staging was 2a (pT3N0Mx).
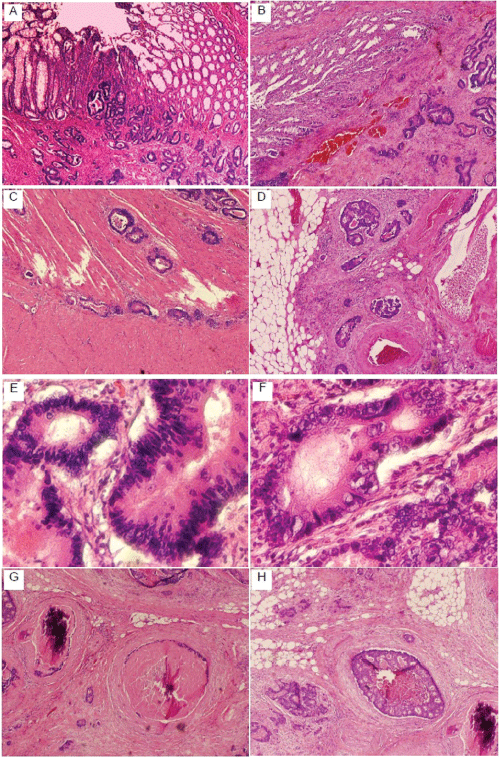
Figures 2: HE staining of sigmoid colon adenocarcinoma. The infiltrating glands of colonic adenocarcinoma originated from mucosa (A) and invade through
submucosa (B), muscular propria (C) and pericolonic fat (D). The neoplastic glands of adenocarcinoma have crowded nuclei with hyperchromatism (E) and
pleomorphism (F). There are also calcifications, necrosis (G) and infiltration of vascular structures (H).
View Figures 2
Discussion
Histopathology type and liver metastasis
It is suggested that pathology features of colon rectal cancer are strongly related to liver metastasis [1]. 46% colon cancer with liver metastasis had morphology of "focal dedifferentiation" which describes polygonal, not columnar, cancer cells in primary tumor. This morphology is considered as a good marker for assessing the tendency of colorectal cancer metastasis to liver [1]. Stromal and perivascular fibrosis extending towards the serosa or adventitia indicates tumor invasion. Lymph node metastasis is independent indicators of liver metastasis [2]. Adenocarcinoma predominantly metastasized to the liver while mucinous adenocarcinoma and signet-ring cell carcinoma more frequently had peritoneal metastasis [3]. This case demonstrated moderately differentiated, high nuclear grade pathology features with pericolonic fat and vascular invasion (T3 stage) in primary tumor. These features are consistent with previous report which showed that in cases of colorectal liver metastasis, 91% are T3 stage and 90% are moderately differentiated in primary tumor [4]. Although abdominal CT scan is routine examination for colon cancer patients and this patient demonstrated liver metastasis in CT scan preoperatively, pathology features of primary tumor and lymph node examination may provide useful information for surgical strategy and prognosis.
Alcohol is one of risk factors for liver metastasis in patients with colorectal carcinoma
Drinking alcohol increases the risk of cancers in gastrointestinal tract. It is reported that lifetime alcohol intake is significantly positively associated with colorectal cancer risk, with higher cancer risks observed in the rectum than distal colon and proximal colon [5]. When assessed by alcoholic beverages at baseline, the colorectal cancer risk for beer was higher than wine [5]. Liver is the most common site of colon cancer metastasis due to drainage of mesenteric veins into hepatic portal vein. Besides anatomy, alcohol consumption is an independent risk factor for liver metastasis in colorectal carcinoma patients [6]. Alcohol is metabolized by alcohol dehydrogenase in hepatic cells and alcohol at physiologically relevant concentrations can disrupt the endothelial monolayer integrity, which is associated with an increased invasion of cancer cells through the endothelial monolayer [7]. The patient in this case has history of alcohol abuse which presented positive association of alcohol consumption with colon cancer liver metastasis. Patients with colorectal carcinoma who drink alcohol require intensive examination and follow-up with respect to liver metastasis.
Epigenetics of colon cancer and its roles in clinical diagnosis, treatment and prognosis
Epigenetics defines heritable changes in gene expression without changing the DNA sequence. It is regulated by three mechanisms: DNA methylation, histone modifications and microRNA expression [8]. Epigenetic factors are found to play important roles in colon cancer occurrence, metastasis and survival. Results from epigenetic studies are significant for clinical diagnosis, therapy and prognosis in colon cancer patients.
Currently, the most commonly used screening methods in clinic are colonoscopy and guiac-based fecal occult blood tests/fecal immunochemical tests. Noninvasive approach, such as detection of genetic and epigenetic alterations in tumor-specific DNA that is shed into serum or plasma is promising and convenient. DNA Methylation at position 5 of the cytosine ring in a CpG island is initiated and maintained by a set of DNA methyl transferases. Tumor suppressor genes are often methylated and silenced in tumorigenesis [9]. A number of methylation biomarkers have been found in circulating cell-free DNA in blood [10]. The septin 9 gene (SEPT9) codes for a GTP-binding protein associated with filamentous structures and cytoskeleton formation [11]. A recently licensed DNA methylation assay is ColoVantage�, which is blood-based and operates based on methylation of the SEPT9 gene [12]. The overall sensitivity of the SEPT9 methylated DNA assay was found to be 90%, with sensitivity for early stage colorectal cancer (stages 1 and 2) diagnosis being 87% and late stage diagnosis (stages 3 and 4) 100%. Colorectal cancer was detected in the cecum, the rectum, and the sigmoid colon in most cases by using this assay. Other assays focusing on SEPT9 methylation that are currently used for the detection of colon cancer are Epi proColon� 1.0 and Abbott Real Time mS9 [13]. ColoSure� test is a fecal-based methylation assay that is clinically available and focuses on methylation of the vimentin gene [14]. This test is recommended for use along with colonoscopy. A sensitivity ranging from 38-88% is suggested for this assay. Runt-related transcription factor 3 (RUNX3) is a serum-based marker [15]. Its promoter�s hypermethylation could potentially be used in the diagnosis of colorectal cancer in CpG island methylator phenotype and microsatellite instability plus CpG island methylator phenotype. Diagnostic panels are being studied. NKX2-5, SPOCK2, SLC16A12, DPYS and GALR2 are candidate biomarkers for colon cancer (methylation range 60%-95%) and GALR2 hypermethylation showed a sensitivity of 85% and specificity of 95% [16]. A DNA methylation correlation network showed cancer specific genes in seven cancers. High-risk group and low-risk group were distinguished by eight biomarkers (ZBTB32, OR51B4, CCL8, TMEFF2, SALL3, GPSM1, MAGEA8, and SALL1) in colon cancer, which provided reference for individual treatment [17]. A 7-gene panel (ANXA3, CLEC4D, LMNB1, PRRG4, TNFAIP6, VNN1 and IL2RB) discriminated colon cancer in the training set. This blood-based biomarker panel can stratify subjects according to their current relative risk across a broad range in an average-risk population [18]. Deleted in liver cancer 1 (DLC1) is a tumor suppressor gene. Detection of DLC-1 methylation in serum would be more useful in prognosis [19]. Neurogenin1 (NEUROG1) gene methylation is a serum marker that has a higher sensitivity than SEPT9 and vimentin for colorectal cancers at early stage. It would be particularly important in diagnosis of asymptomatic colon cancer of any stage [20].
Epithelial�to-mesenchymal transition (EMT) is a fundamental process in development and tumor progression [21]. MicroRNAs are highly conserved ~22-nucleotides single-stranded RNAs that can regulate expression of target genes [22]. It is suggested that microRNAs modulate EMT and facilitate cancer metastasis [23-25] (Table 1). Mir-21, mir-93 and mir-31 were found to be up regulated in liver metastasis [26-28]. Mir-21 and mir-31 are associated with poor prognosis and advanced tumor stage [29]. miR-29a is a potential colon cancer diagnostic marker. It was demonstrated that plasma miR-29a could distinguish colon cancer and advanced adenoma from normal group with sensitivities of 69 and 62.2%, and specificities of 89.1 and 84.7%, respectively [29]. Additionally, serum mir-29a level was significantly higher in colorectal cancer with liver metastasis patients than in colorectal cancer patients. This enabled differentiation between metastatic and non-metastatic tumors [30]. Double cortin-like kinase 1 (DCLK1) promoter is hypermethylated in the vast majority of colorectal cancers (134/164; 82%), with no methylation in the normal mucosa samples (0/106) [31]. DCLK1 is a cancer stem cell specific biomarker and targeting the DCLK1-postive cancer cells is a potential therapy for colorectal cancer.
Molecular genetics of colon cancer and its application in colon cancer metastasis therapy
Somatic mutations play an important role in colon cancer development and metastasis. Molecular genetics studies in this aspect are important in identifying molecular targets of colon cancer. This includes at least two important cell surface receptors: the epidermal growth factor receptor (EGFR) and the Vascular Endothelial Growth Factor Receptor (VEGFR). EGFR is often up-regulated in colorectal cancer while activation of VEGFRs plays an essential role in cancer progression [32,33]. Two groups of agents targeting either VEGFR or EGFR have been added to the therapeutic regimen against metastatic colorectal cancer. Available agents in these groups are the anti-VEGF antibody bevacizumab and the anti-EGFR antibodies cetuximab and panitumumab. They are usually given along with 5-Fluorouracil (5-FU) for metastatic colorectal cancer treatment [34,35].
K-ras mutations are important in the initiation of adenocarcinoma [36]. Approximately 40% of colorectal cancers are characterized by KRAS mutations in codon 12 and 13 in exon 2. These patients have poor prognosis and have increased risk of developing liver and lung metastasis [37-40]. KRAS mutation is negative predictive biomarkers for anti-EGFR therapy among patients with advanced colorectal cancer, including colon cancer with liver metastasis [41]. Array-based tests have been established for profiling of the KRAS mutation status on clinical tumor samples. The chip-based DNA hybridization technique seems to be a promising tool for KRAS mutation analysis to further improve targeted cancer treatment [42].
The serine-threonine kinase, v-raf murine sarcoma viral oncogene homolog B (BRAF), is the principal effector of KRAS. Mutations in BRAF are found in 10 % of colorectal cancer and BRAF mutant metastatic colorectal cancer appears to be a discrete disease subtype with a distinct patient population and significantly poorer survival [43,44]. Single substitution missense mutation V600E that occurs in the kinase domain of the gene is the most common oncogenic mutation and accounts for more than 95% of the BRAF mutations in colorectal cancer [45]. The BRAFV600E in colorectal is associated with advanced cancer stage, poor differentiation, mucinous histology, microsatellite instability, CpG island methylator phenotype [46]. BRAF wild-type is required for response to panitumumab or cetuximab and could be used to select patients who are eligible for the treatment [47]. A BRAFV600E inhibitor, vemurafenib, is being investigated and demonstrated selective sensitivity of colorectal cancer cell lines and xenografts harboring BRAFV600E mutation [47,48].
Summary
Incidence of late stage colorectal cancer ranged from 51.0% to 86.5%. Liver is dominant dissemination site and hepatic metastases develop in approximately half of colorectal cancer cases. Only a small portion of these patients are resectable at the time of diagnosis. Risk factors for colon cancer liver metastasis include age, life style, and pathological character of primary tumor, genetic and epigenetic profiles. Investigate morphology and mechanisms of colon cancer cell dissemination to liver are critical for early diagnosis, prompt treatment and prevention. Epigenetic factors are being used as noninvasive method in clinical diagnosis. Results from molecular genetics studies on colon cancer are being applied in adjuvant chemotherapy in metastasis. Genetics and epigenetics will be important for early diagnosis, therapy and prognosis in cancer patients.
References-
Ono M, Sakamoto M, Ino Y, Moriya Y, Sugihara K, et al. (1996) Cancer cell morphology at the invasive front and expression of cell adhesion-related carbohydrate in the primary lesion of patients with colorectal carcinoma with liver metastasis. Cancer 78: 1179-1186.
-
Inomata M, Ochiai A, Sugihara K, Moriya Y, Yamaguchi N, et al. (1998) Macroscopic features at the deepest site of tumor penetration predicting liver metastases of colorectal cancer. Jpn J Clin Oncol 28: 123-128.
-
Hugen N, van de Velde CJ, de Wilt JH, Nagtegaal ID (2014) Metastatic pattern in colorectal cancer is strongly influenced by histological subtype. Ann Oncol 25: 651-657.
-
Park MS, Yi NJ, Son SY, You T, Suh SW, et al. (2014) Histopathologic factors affecting tumor recurrence after hepatic resection in colorectal liver metastases. Ann Surg Treat Res 87: 14-21.
-
Ferrari P, Jenab M, Norat T, Moskal A, Slimani N, et al. (2007) Lifetime and baseline alcohol intake and risk of colon and rectal cancers in the European prospective investigation into cancer and nutrition (EPIC). Int J Cancer 121: 2065-2072.
-
Maeda M, Nagawa H, Maeda T, Koike H, Kasai H (1998) Alcohol consumption enhances liver metastasis in colorectal carcinoma patients. Cancer 83: 1483-1488.
-
Xu M, Chen G, Fu W, Liao M, Frank JA, et al. (2012) Ethanol disrupts vascular endothelial barrier: implication in cancer metastasis. Toxicol Sci 127: 42-53.
-
Egger G, Liang G, Aparicio A, Jones PA (2004) Epigenetics in human disease and prospects for epigenetic therapy. Nature 429: 457-463.
-
Laird PW, Jaenisch R (1994) DNA methylation and cancer. Hum Mol Genet 3 Spec No: 1487-1495.
-
Schwarzenbach H, Hoon DS, Pantel K (2011) Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 11: 426-437.
-
Hall PA, Russell SE (2004) The pathobiology of the septin gene family. J Pathol 204: 489-505.
-
Warren JD, Xiong W, Bunker AM, Vaughn CP, Furtado LV, et al. (2011) Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med 9: 133.
-
Toth K, Sipos F, Kalmar A, Patai AV, Wichmann B, et al. (2012) Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. PLoS One 7: e46000.
-
Ned RM, Melillo S, Marrone M (2011) Fecal DNA testing for Colorectal Cancer Screening: the ColoSure� test. PLoS Curr 3: RRN1220.
-
Nishio M, Sakakura C, Nagata T, Komiyama S, Miyashita A, et al. (2010) RUNX3 promoter methylation in colorectal cancer: its relationship with microsatellite instability and its suitability as a novel serum tumor marker. Anticancer Res 30: 2673�2682.
-
Chung W, Kwabi-Addo B, Ittmann M, Jelinek J, Shen L, et al. (2008) Identification of novel tumor markers in prostate, colon and breast cancer by unbiased methylation profiling. PLoS One 3: e2079.
-
Zhang C, Zhao H, Li J, Liu H, Wang F, et al. (2015) The Identification of Specific Methylation Patterns across Different Cancers. PLoS One 10: e0120361.
-
Marshall KW, Mohr S, Khettabi FE, Nossova N, Chao S, et al. (2010) A blood-based biomarker panel for stratifying current risk for colorectal cancer. Int J Cancer 126: 1177-1186.
-
Wu PP, Zou JH, Tang RN, Yao Y, You CZ (2011) Detection and Clinical Significance of DLC1 Gene Methylation in Serum DNA from Colorectal Cancer Patients. Chin J Cancer Res 23: 283-287.
-
Herbst A, Rahmig K, Stieber P, Philipp A, Jung A, et al. (2011) Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am J Gastroenterol 106: 1110-1118.
-
Ma L, Lu MF, Schwartz RJ, Martin JF (2005) Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development 132: 5601-5611.
-
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281-297.
-
Bouyssou JM, Manier S2, Huynh D2, Issa S3, Roccaro AM2, et al. (2014) Regulation of microRNAs in cancer metastasis. Biochim Biophys Acta 1845: 255-265.
-
White NM, Fatoohi E, Metias M, Jung K, Stephan C, et al. (2011) Metastamirs: a stepping stone towards improved cancer management. Nat Rev Clin Oncol 8: 75-84.
-
Zhang J, Ma L (2012) MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev 31: 653-662.
-
Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, et al. (2008) MicroRNA-21(miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27: 2128�2136.
-
Fang L, Deng Z, Shatseva T, Yang J, Peng C, et al. (2011) MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin- �8. Oncogene 30: 806-821.
-
Drusco A, Nuovo GJ1, Zanesi N1, Di Leva G1, Pichiorri F1, et al. (2014) MicroRNA profiles discriminate among colon cancer metastasis. PLoS One 9: e96670.
-
Dong Y, Wu WK, Wu CW, Sung JJ, Yu J, et al. (2011) MicroRNA dysregulation in colorectal cancer: a clinical perspective. Br J Cancer 104: 893-898.
-
Wang LG, Gu J (2012) Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol 36: e61-67.
-
Vedeld HM, Skotheim RI, Lothe RA, Lind GE (2014) The recently suggested intestinal cancer stem cell marker DCLK1 is an epigenetic biomarker for colorectal cancer. Epigenetics 9: 346-350.
-
Personeni N, Fieuws S, Piessevaux H, De Hertogh G, De Schutter J, et al. (2008) Clinical usefulness of EGFR gene copy number as a predictive marker in colorectal cancer patients treated with cetuximab: a fluorescent in situ hybridization study. Clin Cancer Res 14: 5869�5876.
-
Jayasinghe C, Simiantonaki N, Kirkpatrick CJ (2015) Cell type- and tumor zone-specific expression of pVEGFR-1 and its ligands influence colon cancer metastasis. BMC Cancer 15: 1130.
-
Meyerhardt JA, Mayer RJ (2005) Systemic therapy for colorectal cancer. N Engl J Med 352: 476-487.
-
Feng QY, Wei Y, Chen JW, Chang WJ, Ye LC, et al. (2014) Anti-EGFR and anti-VEGF agents: important targeted therapies of colorectal liver metastases. World J Gastroenterol 20: 4263-4275.
-
Iwanaga K, Yang Y, Raso MG, Ma L, Hanna AE, et al. (2008) Pten inactivation accelerates oncogenic K-ras-initiated tumorigenesis in a mouse model of lung cancer. Cancer Res 68: 1119-1127.
-
Kemeny NE, Chou JF, Capanu M, Gewirtz AN, Cercek A, et al. (2014) KRAS mutation influences recurrence patterns in patients undergoing hepatic resection of colorectal metastases. Cancer 120: 3965-3971
-
Karagkounis G, Torbenson MS, Daniel HD, Azad NS, Diaz LA Jr, et al. (2013) Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer 119: 4137-4144.
-
Vauthey JN, Zimmitti G, Kopetz SE, Shindoh J, Chen SS, et al. (2013) RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg 258: 619-626.
-
Stremitzer S, Stift J, Gruenberger B, Tamandl D, Aschacher T, et al. (2012) KRAS status and outcome of liver resection after neoadjuvant chemotherapy including bevacizumab. Br J Surg 99: 1575-1582.
-
De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, et al. (2010) Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 304: 1812-1820.
-
Steinbach C, Steinbr�cker C, Pollok S, Walther K, Clement JH, et al. (2015) KRAS mutation screening by chip-based DNA hybridization - a further step towards personalized oncology. Analyst 140: 2747-2754.
-
Davies H, Bignell GR, Cox C, Stephens P, Edkins S, et al. (2002) Mutations of the BRAF gene in human cancer. Nature 417: 949-954.
-
Tol J, Nagtegaal ID, Punt CJ (2009) BRAF mutation in metastatic colorectal cancer. N Engl J Med 361: 98-99.
-
Saridaki Z, Tzardi M, Sfakianaki M, Papadaki C, Voutsina A, et al. (2013) BRAFV600E mutation analysis in patients with metastatic colorectal cancer (mCRC) in daily clinical practice: correlations with clinical characteristics, and its impact on patients' outcome. PLoS One 8: e84604.
-
Farina-Sarasqueta A1, van Lijnschoten G, Moerland E, Creemers GJ, Lemmens VE, et al. (2010) The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol 21: 2396-2402.
-
Tie J, Desai J (2014) Targeting BRAF mutant metastatic colorectal cancer: clinical implications and emerging therapeutic strategies. Target Oncol .
-
Hirschi B, Gallmeier E, Ziesch A, Marschall M, Kolligs FT (2014) Genetic targeting of B-RafV600E affects survival and proliferation and identifies selective agents against BRAF-mutant colorectal cancer cells. Mol Cancer 13: 122.





