International Journal of Brain Disorders and Treatment
MicroRNA in Brain Neoplasia: A Review
Michela Visani1, Giorgia Acquaviva2, Gianluca Marucci3, Moira Ragazzi4, Enrico Fraceschi*5, Alba A Brandes5, Giovanni Tallini2, Annalisa Pession1 and Dario de Biase2*
1Department of Pharmacology and Biotechnology (FaBiT), University of Bologna, Italy
2Department of Medicine (DIMES), University of Bologna, Bellaria Hospital, Italy
3Department of Biomedical and NeuroMotor Sciences (DiBiNeM), Bellaria Hospital, University of Bologna, Italy
4Pathology Unit, IRCCS-Arcispedale Santa Maria Nuova, Reggio Emilia, Italy
5Medical Oncology Department, Bellaria Hospital, Azienda USL/ IRCCS Institute of Neurological Sciences, Italy
*Corresponding author:
Dario de Biase, BSc, PhD, Department of Medicine, University of Bologna, Molecular Biology Lab - Anatomic Pathology Bellaria Hospital, via Altura 3, 40139 Bologna, Italy, Tel: 39-051-622-5752, Fax: 39-051-622-5759, E-mail: dario.debiase@unibo.it
Int J Brain Disord Treat, IJBDT-1-002, (Volume 1, Issue 1), Review Article; ISSN: 2469-5866
Received: July 30, 2015 | Accepted: August 25, 2015 | Published: August 27, 2015
Citation: Visani M, Acquaviva G, Marucci G, Ragazzi M, Fraceschi E, et al. (2015) MicroRNA in Brain Neoplasia: A Review. Int J Brain Disord Treat 1:002. 10.23937/2469-5866/1510002
Copyright: © 2015 Visani M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
MicroRNAs (miRNAs) are small RNAs involved in regulation of several cellular processes and are involved in the silencing of cell's message in a variety of ways. In cancer, miRNAs can be involved in the regulation of important genes involved in tumorigenesis, tumor development, and angiogenesis. For these reasons, miRNAs could have considered oncogenic-miR (miRNA with oncogenic roles) or oncosuppressor-miR (miRNA with tumor suppressor roles). MiRNAs may alter the expression of genes involved in cell cycle, regulation of cell proliferation, apoptosis, angiogenesis, immune response, tumor invasion and metastasis, and genomic instability of cancer cells. Deregulation of miRNA expression has been demonstrated in each cancer type studied (e.g. breast and ovarian carcinomas, non-small cell lung cancer, pancreatic cancer and brain tumors). Moreover, miRNA expression signatures could be associated with well-defined clinicopathologic features and disease outcome and prognosis. This review focuses on the studies from 2006 to 2013 investigating miRNAs differentially expressed in different type of brain tumors.
Keywords
MicroRNA, Glioblastoma, Medulloblastoma, Brain neoplasia, Review
Introduction
MicroRNAs (miRNAs) are small RNAs, about 21 nucleotides in length (19-23 nucleotides), involved in regulation of proliferation, differentiation, and apoptosis during normal development. MiRNAs are involved in the silencing of the cell's own message in a variety of ways, as translational repression and mRNA cleavage (Figure 1) [1-3]. MiRNAs may also act by fine-tuning target activity, simultaneously modulating multiple targets and deregulating targets for rapid reactivation. More than a thousands of annotated miRNA sequences in human genome are available in the latest update of miRBase (June 2013, http://www.mirbase.org/) and they are thought to regulate more than one third of human genes [4,5].
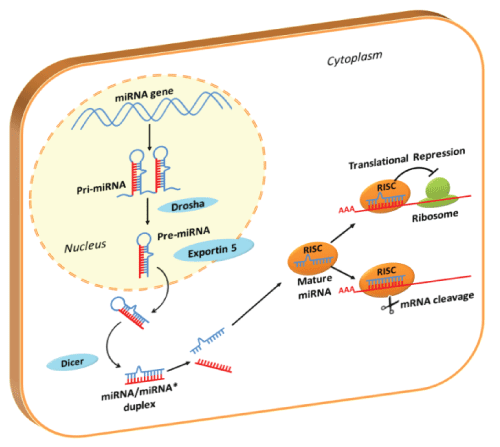
.
Figure 1: Pathway of Translational Silencing by miRNAs
In the nucleus miRNAs sequences are transcribed by RNA polymerase (mainly RNA polymerase II): the long, stem-loop primary transcript (pri-miRNA) is further processed in ~60-70nt pre-miRNA by nuclear Drosha RNase III endonuclease. Pre-miRNAs are transported into cytoplasm by Exportin-5 system: once in the cytoplasm, Dicer RNase III endonuclease, process miRNA precursors into mature 19-23nt miRNA duplexes. These duplexes comprise the mature miRNA and complementary fragment derived from the opposing arm of the pre-miRNA (miRNA*). Only the strand with the less stable hydrogen bonding at its 5'-end is selected (mature miRNA), while the complementary miRNA* is degraded. Mature miRNA is incorporated into ribonucleoprotein complexes known as RNA-induced silencing complexes (RISC). The major components of miRNPs are proteins of the Argonaute family: these proteins play a central role in binding the mature miRNA and drive it to mRNAs target recognition.
MiRNA-RISC complexes recognize and link miRNA complementary regions in 3'-UTR of target mRNAs. According to the grade of homology with the mRNAs, miRNAs could act in two ways: a perfect complementarity leads to mRNA cleavage, while an imperfect complementarity represses mRNA translation. RISC: RNA-Induced Silencing Complex.
View Figure 1
Aberrant miRNA expression has been demonstrated in each cancer type studied [6], as breast cancer [7], ovarian carcinomas [8], non-small cell lung cancer [9] or pancreatic cancer [10].
Brain tumors are one of the top 10 causes of cancer deaths and an early and accurate diagnosis is essential for disease management. Brain tumors could be classified into glial tumors (e.g. glioblastoma, astrocytomas, oligodendroglial tumors, ependymal tumors), embryonal tumors (e.g. medulloblastomas), tumors of the the meninges (e.g. meningiomas), tumors of the hematopoietic system and tumor of the sellar region [11]. Introduction of molecular markers as 1p/19q codeletion [12], O6-methylguanine methyltransferase (MGMT) methylation status [13] or isocitrate dehydrogenase (IDH1 or IDH2) mutations [14] have provided new therapeutic or prognostic tools but other molecules are needed to improve management of these tumors. From here, it is gathered the necessity to discover new molecules as potentially useful parameters for prognosis, diagnosis or target therapy strategies and miRNAs could represent important molecular biomarkers.
This review focuses on the studies investigating different miRNAs expression profiles in different type of brain tumors from January 2006 to June 2013.
Starting Material and Techniques
Starting material
Cell Lines: Cell lines are one of the most used starting materials for expression analysis. This is due to the evidence that extracting RNA from cell lines allows to obtain a good quality and quantity of the nucleic acid. However, considering that miRNAs are more resistant than mRNA to degradation, the need of high-quality RNA is less crucial for miRNAs analysis. Moreover, it should be considered that miRNA expression is highly variable and results obtained in cell lines, in particular from immortalized ones, could provide results far away from those obtained in "ex vivo" samples. This could be due to changes in cultured cells, but also to the fact that cell lines are not influenced by microenvironment as happened in ex vivo specimens. Several analysis of miRNA expression in brain cell lines have been performed [15-20], however these results should be confirmed in ex vivo specimens for determining brain tumor miRNAs profile (Figure 2).
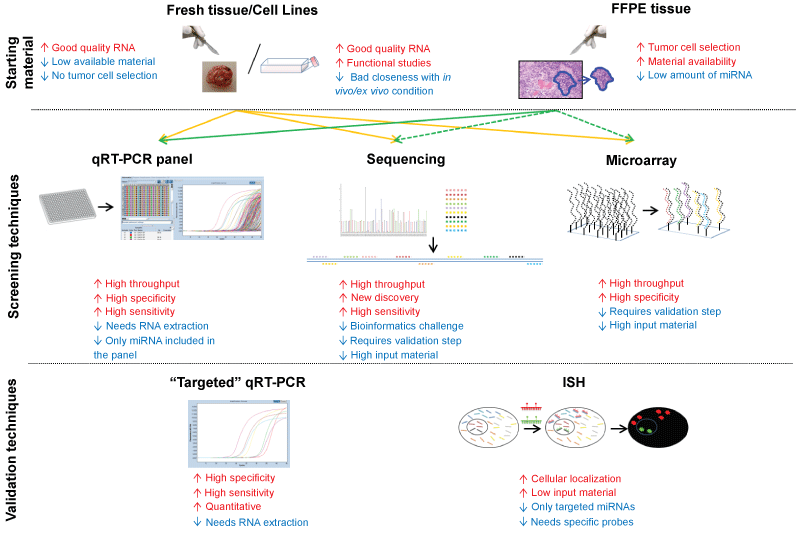
.
Figure 2: Scheme of possible sources of starting material for miRNA analysis and main available techniques. Continues arrows mean that material is optimal for the techniques. Dotted arrows indicate that material is not recommended for the technique, due to high input of miRNA required by the assay. qRT-PCR: quantitative Real-Time PCR, ISH: In Situ Hybridization.
View Figure 2
"Fresh/Frozen" specimens: The limitation of "fresh" brain tissue for analyzing miRNAs as biomarkers is the extremely invasive procedure for collecting material. In fact, if on the one hand non-formalin fixed specimens allow obtaining a huge amount of miRNA, on the other their availability from surgical samples is limited, considering that material for diagnostic evaluation needs to be preserved. For this reason, the analysis of miRNAs starting from formalin-fixed paraffin-embedded (FFPE) specimens could provide greater amount of material for research purposes. Due to the good amount of good quality material obtained from fresh/frozen specimens, several papers have investigated miRNAs expression starting form this type of samples [15,21-56] (Figure 2).
FFPE samples: Starting from formalin fixed and paraffin embedded (FFPE) samples might be of great usefulness for miRNAs expression studies. Due to their short length, the mature miRNAs seem not to be influenced by nucleic acid degradation caused by formalin fixation [57], as happened on the contrary for long RNA or DNA. Some papers reported the feasibility of miRNAs expression from FFPE specimens in different tissues as kidney [58], prostate [59] or breast [60]. Several studies have analyzed miRNA expression profile in FFPE specimens compared with non-neoplastic or benign tissue, obtaining robust and reliable results [26,37,61-66]. It should be considered that: using FFPE samples, a neoplastic cells enrichment step (micro or macro-dissection) was feasible; FFPE samples could be easily retrieved from the archives of anatomic pathology institutes and, furthermore, the results displayed a good correlation of miRNA expression values obtained in FFPE specimens and in the corresponding fresh/frozen material [26,67](Figure 2) .
Another important aspect to consider in miRNA expression analysis is the selection of control group. There are different specimens that could be used as normal reference for miRNAs analysis in brain samples as the normal area adjacent the tumor, a commercial reference or cell lines. As previously demonstrated by our group, the selection of control group is a crucial step in miRNA analysis; in fact, it is possible to obtain discrepant results in miRNA expression data simply due to different choice of non-neoplastic control [68].
Techniques
Analysis of miRNAs with high specificity and sensitivity is technically challenging as mature miRNAs are short molecules, differ in GC (guanine-cytosine) content, and with single nucleotide difference within miRNA families. Several techniques based on extracted RNA (e.g. deep sequencing, microarrays, quantitative Real-Time Polymerase Chain Reaction - qRT-PCR), or in situ hybridization assays (ISH) have made possible to profile miRNAs in brain lesions with high accuracy (Figure 2).
Microarray: Microarray analysis is a high-throughput assay that allows monitoring the expression of thousands of miRNAs in a single experiment. In the detection of miRNAs by microarray, appropriate probe design is crucial. In fact, capture probes must have a high specificity and affinity for individual transcripts. Because miRNAs are small molecules, the probe design possibilities are limited on the miRNA sequence. The microarray techniques should not be used to make quantitative statements, but rather to determine the relative change in expression between two states (e.g. tumoral vs. non-tumoral) [69]. Moreover, it should be considered that another limitation of microarray analysis is that the results need to be validated using a targeted technique, as qRT-PCR or ISH. Several papers have performed a genome-wide microarray analysis of miRNA expression of neoplastic brain specimens to detect expression signatures and associate this profile with characterization of the tumor [15-18,20,22,24,27,28,31,34,35,37,39,41,43-45,49,50,55,62,63] (Figure 2).
Next Generation Sequencing: Next generation sequencing (NGS) platforms became available for the sequencing sRNAs, including miRNAs, and allows to analyze thousands sequence in parallel from a given sample. Moreover NGS could highlight both miRNAs present in few copies and discover novel miRNAs, without the limitation of microarray probe selection. The high sensitivity of NGS offers an advantage over microarray assays, because the high variability of miRNA expression complicates the detection of miRNAs expressed at low copy numbers. However, because each NGS experiment produces several giga base pairs (Gbp) of sequence data, this technique needs massive bioinformatics challenges to analyze and handle sequence information. Due to relative novelty of this approach and to the bioinformatics challenges to analyze the output data, NGS has been used less than other techniques for miRNAs analysis in brain neoplasia (Figure 2) [17,19,33,52].
qRT-PCR: Although high throughput assays (e.g. microarray or NGS) are useful tools to provide a broad overview of the miRNA expression, these data need to be confirmed by "miRNA specific" approaches. Quantitative real-time reverse transcriptase polymerase chain reaction is a method frequently used either to assessing expression in brain neoplasia of specific miRNAs and to confirm data obtained with broad range techniques [21,23,25,26,29,30,32,36-38,40,42,45-47,51-54,56,61,64-66]. Several approaches to analyze miRNAs continue to be based on single-target assays; however, high-throughput qRT-PCR technologies can be employed to identify differences in miRNA expression levels. For example, TaqMan low density arrays (TLDA) and Locked Nucleic Acid (LNA) array platform have been introduced for miRNA detection (Figure 2).
In situ Hybridization (ISH): A limitation of previously described techniques is that they are based on extracted miRNA and then do not provide information regarding the cell type present in the sample and the specific site of expression of a particular miRNA. Thus, the use of an ISH/IHC assay (in situ hybridization/immunohistochemistry) allows rapid and direct evaluation of changes of miRNA expression within the lesional cells. In situ techniques are used either as main assay for evaluating miRNA expression in pancreatic lesions visualized at cellular/subcellular level or to confirm results obtained by qRT-PCR or high-throughput technique (e.g. miRNA microarray or real-time array, see above) (Figure 2) [62,64].
MiRNA Expression in Glioblastoma
Glioblastoma multiforme (GBM) is the most malignant and the most frequent brain tumor of the adult, accounting for about 50% of diffuse gliomas [11,70]. It is a pleomorphic astrocytic tumor, characterized by poor cell differentiation, high cell density, marked nuclear atypia and high mitotic activity. Moreover, microvascular proliferation and necrosis are peculiar features, in association with a high proliferation rate, marked invasiveness and resistance to conventional therapies [11,70]. Despite progresses in surgical techniques, radiotherapy, chemotherapy and "target therapy", the prognosis of this neoplasia remains poor [70]. In the last decades several studies have been performed in attempt to identify a specific miRNAs expression pattern of GBM and a small subset of consistently deregulated miRNAs were further functionally characterized for their activities and downstream targets possibly involved in this tumor [34,44,71-74].
Twenty-four profiling reports have been selected and reviewed for this paper to underline miRNAs found consistently deregulated in GBM samples or in GBM cell lines (Table 1 and Supplementary Table). Among them, 16 studies have identified multiple miRNAs that are differentially expressed in GBM compared to normal tissues [15,24,25,28,31,33,34,37,44,47-50,55,65,66]. Three reports have focused on miRNA expression profiles that differentiate GBM (World Health Organization - WHO - grade IV) from Anaplastic Astrocytoma (AA, WHO grade III) or from low grade (WHO grade II or I) gliomas [32,40,43]; in two studies miRNA profile of GBM stem cells was analyzed in comparison with non-stem cells [16,17]; in another one miRNA differences have been analyzed between GBM migrating cells versus non migrating cells [18]; finally, Zhou et al. analyzed differences in miRNA expressions in 5 GBM cell lines and 1 AA cell line, compared to a commercial RNA [20] while Nyazi et al. analyzed miRNAs expression in GBM short term and long term survivors [63]. A great variability among the different studies is remarkable, depending from the technique used (microarray, qRT-PCR assay or next generation sequencing), the type or the number of samples analyzed, the number of target miRNAs studied through the expression results obtained.
![]()
Table 1: MiRNAs profiling studies in glioblastoma
Only miRNAs showed a fold change greater than or equal to two-fold, were considered. GBM: Glioblastoma, AA: Anaplastic Astrocytoma, NB: Normal Brain, ANB: Adjacent Normal Brain, LGA: Low Grade Astrocytoma, LGG: Low Grade Glioma, O: Oligodendroglioma, DA: Diffuse Astrocytoma, NS: Not Specified, NGS: Next Generation Sequencing, TGCA: The Cancer Genome Atlas (TCGA) Datasets, FFPE: Formalin-Fixed Paraffin-Embedded. In bold, miRNAs reported up- or down-regulated in at least 3 studies. § Only the most 10 up- or down-regulated miRNAs are reported, † The authors considered a cut-off fold change ≥ 1.5.
View Table 1
From this comparison, only few miRNAs (up-regulated: miR-21, miR-10b, miR-15b, miR-16, miR-25, miR-92b, miR-93, miR-106b, miR-155, miR-210, miR-17-5p, miR-106a, miR-148a, miR-196b; down-regulated: miR-132, miR-218, miR-124, miR-128a, miR-323, miR-128, miR-7, miR-181b, miR-221, miR-222, miR-31, miR-138, miR-181c, miR-379) shared the same deregulation pattern in at least 3 studies performed on fresh or FFPE tissues (Tables 1-3 and Supplementary Table). Interestingly, miR-221/222, defined as "oncomirs" in GBM and deeply discussed by several works as important miRNAs in GBM pathogenesis [25,75-79], were found either up- [15,25] or down-regulated [48,49,65,66] (miR-221), or only down-regulated (miR-222) [49,65,66] (Tables 1-3 and Supplementary Table). Intriguingly, miR-221 and miR-222 were found up-regulated in all studies that have investigated their role in cellular function (Tables 2,3) [80-83]. This discrepancy could be due to several experimental variables, as choice of "non-neoplastic control" or source of the starting material, as previously reported by Visani and colleagues [84].
![]()
Table 2: MiRNAs reported up-regulated in glioblastoma samples.
Only miRNAs reported in at least 3 profiling reports were considered (for more information see Table 1). Studies performed on cell lines were not considered for miRNA profile. *Number of studies reporting the same regulation pattern for each miRNA (see Table 1). †In this study, authors obtained a down-regulation of selected miRNA. NA, Functional studies are not available in literature.
View Table 2
![]()
Table 3: MiRNAs reported down-regulated in glioblastoma samples
Only miRNAs reported in at least 3 profiling reports were considered (for more information see Table 1). Studies performed on cell lines were not considered for miRNA profile. ^ Five studies were focused on miR-128a. *Number of studies reporting the same regulation pattern for each miRNA (see Table 1). †In these studies, an up-regulation of selected miRNAs was observed. NA, Functional studies are not available in literature
View Table 3
According to literature data (Tables 1-3 and Supplementary Table), it is clear that by now an exhaustive miRNAs profile of GBM is still far to be well defined.
Many other studies were focused only on a single, or on a small subset of miRNAs to analyze their function, genetic regulation and expression in GBM tissues/cell lines. Functional analysis are very important to experimentally validate possible targets of those miRNAs found deregulated in previous studies, clarifying their molecular roles in GBM pathogenesis and giving information above putative prognostic markers or therapeutic targets [16,20,24,31,32,34,44,47,71-74,79-81,83,85-96].
Oncogenic miRNAs in GBM
MiRNAs found consistently up-regulated in at least 3 studies performed on fresh or FFPE tissues are reported in tables 1 and 2 and in Supplementary Table. Several miRNAs found consistently up-regulated in GBM have been intensively investigated with respect to both expression and functionality (Table 2) [20,24,40,44,52,71,77,80-83,97-112]. In particular, miR-21 and miR-10b had been deeply studied in literature. MiR-21 had been found as significantly deregulated in 15 out of 19 profiling studies performed on FFPE or fresh tissues here considered (Tables 1,2 and Supplementary Table). In all cases an over-expression in GBM tissues or in GBM cell lines was reported [15,24,25,32-34,37,40,43,44,47,48,50,65,66]. Several functional studies have investigated miR-21 role in several processes involved in malignancy progression and maintenance of GBM. It was demonstrated that miR-21 acts as anti-apoptotic factor blocking the expression of critical apoptosis-related genes. Chan et al. [24] first reported that the forced down-regulation of miR-21 leads to caspase 3/7 activation and associated apoptotic cell death. Chen et al. [71] and Gaur et al. [98] showed that in vitro and in vivo miR-21 over-expression inhibits the expression of the pro-apoptotic factor PDCD4. MiR-21 also regulates molecules within the p53 and TGFB apoptotic pathways targeting p63 (a homologue of p53), JMY (a p53 activator), TOPORS, TP53BP2 and HNRPK and the receptors TGFBR2/3, respectively [100]. Also DAXX (death associated protein 6), which acts in both pathways stabilizing p53 and mediating TGF-h apoptosis, is regulated by miR-21 [100].
Some of the several mRNA targets controlled by miR-21 are directly associated with the tumor invasiveness. For example, Gabriely et al. [97] reported that miR-21 regulates MMPs and glioma cell invasiveness by directly controlling the MMP inhibitors RECK and TIMP3. Authors showed that the forced inhibition of miR-21 decreases MMP activities both in vitro and in vivo, leading to reduction of glioma cell motility and invasion [97].
Furthermore, miR-21 could control GBM cell growth by maintaining active the telomerase activity, which is fundamental to maintain the DNA integrity of the replicating tumor cells, establishing cell immortality and survival. As suggested by Wang and colleagues [113], miR-21 regulates hTERT (human telomerase reverse transcriptase) expression in a STAT3- (signal transducer and activator of transcription 3) dependent manner and guarantee GBM cell growth.
In a study by Zhou et al. [20] it was demonstrated that the tumor suppressor PTEN could be a direct target of miR-21. A decreased expression of EGFR and Akt activation was also reported in response to an inhibition of miR-21, concurrently with increased apoptosis through decreased levels of the anti-apoptotic Bcl-2. This suggested that the control of miR-21 on cell growth and apoptosis pathways could be mediated by other targets in parallel with PTEN [20].
In the present series, seven studies reported an up-regulation of miR-10b in GBM tissues [15,33,34,44,47,48,50] (Tables 1,2 and Supplementary Table). In 2009, Sasayama et al. showed that the up-regulation of miR-10b was associated with glioma grading and malignancy, according to data obtained on 43 glioma samples (seventeen GBM, six AA, ten low-grade astrocytoma, six oligodendroglioma and four ependymoma) and six glioma cell lines [44]. Furthermore, they showed that miR-10b inhibits the expression of the transcription factor HOXD10 [44]. It was demonstrated that HOXD10 is able to regulate several genes promoting invasion, migration, extracellular matrix remodeling and tumor progression, including α3-integrin, β-integrin, MMP-14 and two molecules, uPAR (urokinase receptor) and RhoC (Ras homolog gene family member C), which expression is correlated with expression levels of miR-10b [44].
Gabriely et al. [114] demonstrated in several glioma cell lines that miR-10b inhibition could block cell cycle progression and in some cases lead to the accumulation of senescent glioma cells. MiR-10b acts as cell cycle regulator modulating in an indirect manner the cyclins B1 and D1 expression levels, whereas the inhibition of miR-10b expression is significantly associated to lower levels of this two factors [114]. Moreover, miR-10b inhibition is strictly associated to an increase activation of caspase-3 and -7 suggesting a direct role of miR-10b on apoptotic pathway [114].
In the same study, Bim, TFAP2C, p16, and p21 were validated as direct targets of miR-10b, confirming its role in cell cycle and apoptosis control. Intriguingly, in this study the regulation of miR-10b reported on HOXD10 by Sasayama et al. [44] was not confirmed [114].
Lin et al. [99] reported that miR-10b directly regulates inhibition of several tumor suppressor genes (FOXO3, CYLD, HOXD10, TP53, PAX6, PTCH1 and NOTCH1) affecting apoptosis, cell invasion and angiogenesis.
Tumor suppressor miRNAs in GBM
MiRNAs found consistently down-regulated in at least 3 studies performed on fresh or FFPE tissues are reported in tables 1 and 3 and in Supplementary Table. Among miRNAs found consistently down-regulated in GBM (Table 3) [31,33,47,72,80-83,101,115-130], miR-128 and miR-218 have been deeply investigated.
MiR-218 was found deregulated in 5 out of the 19 miRNA profiling studies performed on FFPE or fresh tissues here considered. In all cases, a down-regulation of miR-218 was observed in GBM [28,33,47,48,50] (Tables 1,3 and Supplementary Table).
The lack of miR-218 expression leads to an increase of GBM cell invasiveness. In fact, the down-regulation of miR-218 causes an over-expression of IKK-β which promotes the activation of NF-κB/MMP9 signaling pathway [125]. Also the oncogenic transcription factor LEF-1 regulates the expression of MMP9 and Liu et al. [122] demonstrated that LEF-1 is a direct target of miR-218 supporting the thesis that the deregulation of this miRNA is directly correlated to the GBM cell migration and invasion. In another functional study, Xia and colleagues [129] showed that the over-expression of miR-218 in glioma cells induced apoptosis and inhibited cell viability, proliferation and tumorigenicity. ECOP (Epidermal growth factor receptor-coamplified and over-expressed protein), a proteins that belongs to NF-κB signaling pathway associated with apoptotic response, was identified as a direct target of miR-218 [129].
Both miR-128 [37,43,48] and miR-128a [15,47,50,65,66] were reported as down-regulated in eight out of 19 profiling studies performed on FFPE or fresh tissues. The exogenous expression of miR-128 significantly reduced in vitro and in vivo glioma cell proliferation. One of validated miR-128 target is Bmi1, a molecule that regulates the epigenetic gene silencing by chromatin modifications and stem cell renewal [31,131]. MiR-128 promotes GBM cell proliferation by controlling the expression of ARP5 (angiopoietin-related growth factor protein 5) which normally regulates cell regeneration, proliferation and promotes neovascularization; expression of E2F-3a, a transcription factor which plays a crucial role in the control of cell-cycle progression [131], is also controlled by miR-128. The WEE1 tyrosine kinase, that phosphorylates CDK1 playing a role in cell-cycle control, is a direct target of miR-128, as reported by Wutchy et al. [128].
Papagiannakopoulos and colleagues [132] suggested that miR-128 could also control the mitogenic tyrosine kinase signaling targeting EGFR and PDGFRα oncogenes. Finally, Shi et al. [133] reported p70S6K1 as another direct target of miR-128, which is involved in glioma angiogenesis and tumorigenesis.
MiRNA Expression in other Glial tumors
MiRNA expression in Astrocytoma
Diffuse astrocytoma (WHO grade II) is characterized by a high degree of cellular differentiation and slow growth. Diffuse astrocytoma inherently tends to locally recur and spontaneously progress to AA (WHO grade III) and eventually to secondary GBM (WHO grade IV). Progression from primary low-grade glioma to secondary GBM was associated with up-regulation of 12 miRNAs (miR-9, miR-15a, miR-17, miR-19a, miR-20a, miR-21, miR-25, miR-28, miR-130b, miR-140, miR-210) and with the down-regulation of 2 miRNAs (miR-184 and miR-328) [134] (Table 4).
![]()
Table 4: MiRNAs deregulated in astrocytomas
AA: Anaplastic Astrocytomas, GBM: Glioblastoma, qRT-PCR: quantitative Real-Time Polymerase Chain Reaction, NGS: Next Generation Sequencing.
View Table 4
The miR-21 and miR-221 were also observed up-regulated in astrocytoma [25,43,53], a diffuse infiltrating malignant astrocytoma (WHO grade III) characterized by nuclear atypia, increased cellularity and significant mitotic activity. In contrast, miR-106a, miR-124, miR-125 and miR-181b were reported as down-regulated in this type of tumors [25,53].
As regards some of the above reported deregulated miRNAs, miR-181b, miR-106a and miR-21 were seen significantly associated with poor survival in patients with astrocytoma. These data could lead to hypothesize that miR-21, miR-106a or miR-181b may have a strong potential to serve as novel prognostic markers of astrocytoma [53].
The miRNAs analysis in the serum of malignant astrocytoma patients, compared to normal controls, revealed several miRNAs (miR-15b*, miR-23a, miR-133a, miR-150*, miR-197, miR-497 and miR-548b-5p) whose concentrations were significantly lower in serum of the patients. Moreover, the concentrations of miR-23a, miR-150*, miR-197 and miR-548b-5p were markedly decreased in malignant cases if compared to the astrogliosis group [52].
Considering miRNAs found deregulated in at least 2 studies, miR-21 results to be up-regulated in astrocytoma while miR-181b is down-regulated.
MiRNA expression in oligodendroglial tumors
Oligodendrogliomas (WHO grade II) are diffusely infiltrating, well-differentiated gliomas, composed of neoplastic cells morphologically resembling oligodendroglia [11] (Table 5).
![]()
Table 5: MiRNAs deregulated in oligodendroglial tumors
GBM: Glioblastoma, ISH: in situ hybridization, qRT-PCR: quantitative Real-Time Polymerase Chain Reaction
View Table 5
A comparative study between ten oligodendrogliomas and non-neoplastic samples identified several miRNAs up- (let-7a, let-7f, miR-17, miR-21, miR-155, miR-17, miR-16, miR-26b, miR-374a, let-7d, miR-20a, miR-15b, miR-7b, miR-9) or down-regulated (miR-132, miR-134, miR-7, miR-330-3p, miR-127-3p) [37]. Moreover, in the same study, a set of seven markers (miR-21, miR-128, miR-132, miR-134, miR-155, miR-210 and miR-409-5p) was proposed to help in distinguishing GBM from oligodendroglial tumors [37].
Nelson et al. demonstrated that miR-9 was highly expressed in oligodendroglioma and fetal brain, but had a low expression in adult brain, suggesting that miR-9 up-regulation may be important in the development of oligodendrogliomas [62]. The same authors observed reduced level of miR-124 in oligodendroglioma compared to normal brain [62].
A comparison between newly diagnosed anaplastic oligodendroglioma and recurrent tumors showed that 7 miRNAs (miR-124, miR-128, miR-139-5p, miR-153, miR- 210, miR-582-5p and miR-96) were highly increased in the patients with recurrent anaplastic oligodendrogliomas. In particular, 5 miRNAs (miR-124, miR-128, miR-139-5p, miR-210 and miR-582-5p) exhibited more than 10-fold increased expression [35].
In contrast, 21 miRNAs (miR-1, miR-1180, miR-133b, miR-135b, miR-1539, miR-193a-5p, miR-196a, miR-196b, miR-200b, miR-21*, miR-221*, miR-224, miR-24-1*, miR-31, miR-31*, miR-32*, miR-34a*, miR-34c-5p, miR-455-5p, miR-503 and miR-631) showed a huge (0.01-fold) decreased expression in patient with recurrent anaplastic oligodendroglioma, if compared with those with newly diagnosed tumors [35].
Moreover, in oligodendroglioma also miR-137 was found down-regulated, as already reported for oligoastrocytoma [38]. MiR-9 is the only miRNA found deregulated (up-regulated) in oligodendroglioma in at least 2 papers [37,62].
OligoAstrocytomas. Oligoastrocytomas are diffuse infiltrating glioma composed of a mixture of two distinct neoplastic cell types morphologically resembling the tumor cells in oligodendroglioma and diffuse astrocytoma (WHO Grade II) [11].
In a study of 16 oligoastrocytomas, miR-137 was down-regulated in the vast majority of analyzed samples [38]. According to data obtained in transfected cell lines, miR-137 could play an important role in the suppression of invasion of glioma cells. A putative target of miR-137 in oligoastrocytomas could be CSE1L (Chromosome Segregation 1-Like), a protein involved in tumor cell invasion and metastasis in cancer progression [38].
MiRNA expression in ependymal tumors
Ependymoma (WHO grade II) is a common pediatric central nervous system (CNS) tumor that is believed to originate from ependymal cells located in the lining of ventricular surfaces in the brain [11] (Table 6).
![]()
Table 6: MiRNAs deregulated in ependymoma
qRT-PCR: quantitative Real-Time Polymerase Chain Reaction
View Table 6
Costa and colleagues analyzed the expression of 365 miRNAs in 34 ependymoma samples compared with normal brain tissue of the same patients [26]. The authors identified 28 miRNAs that were differentially expressed in ependymoma samples. Specifically, 23 miRNAs (miR-10a, miR-17-5p, miR-19a, miR-19b, miR-20a, miR-21, miR-32, miR-34a, miR-106b, miR-130a, miR-135a, miR-142-3p, miR-193a, miR-210, miR-301, miR-449b, miR-502, miR-518b, miR-551b, miR-565, miR-591, miR-594 and miR-601) were up-regulated and 5 miRNAs (miR-139, miR-323, miR-383, miR-433 and miR-485-5p) were down-regulated in ependymoma specimens [26]. Moreover, miR-203 was proposed as a biomarker for management of disease progression in ependymomas and as a predictor for the likelihood of recurrence. In fact, while higher expression of miR-432, miR-411, miR-376a, miR-381 and miR-487b was associated with a lower relapse free probability, a lower expression of miR-203 was correlated to a trend to develop recurrences [26].
MiRNA Expression in Medulloblastoma
Medulloblastoma (MB, WHO grade IV), the most frequent brain neoplasia in children, is an aggressive infratentorial primitive neuroectodermal tumor that originates from aberrant development of cerebellar progenitor neurons [11,135,136]. Nevertheless, the division into molecular subgroups is a useful tool for the risk stratification of the patients and has generated several hypotheses for new treatment [137]; further molecular characterization will allow the identification of novel targeted therapies and will improve the reliability of risk stratification. To develop novel therapies and to identify new biomarkers useful to classify histological variants or to predict patient's overall survival, in the last years many groups have investigated the miRNAs layout of this neoplasia [138]. In table 7, six miRNA profiling studies are reported: miRNAs expression in MB samples has been evaluated, for example, in fresh tumors compared with non-neoplastic samples [29,39,42] or with CD133+ Neuronal Stem Cells (NSCs) [30] or, as in the study of Uziel et al., a rodent model of MB was studied [19]. In the work of Ferretti et al. [56] they compared miRNA expression patterns between MB with high or normal expression of Gli1, a transcription factor of Sony Hedgehog (SHH) signaling pathway which is often hyperactive in MBs [56,135]. They obtained 31 significantly deregulated miRNAs, most of them were down-regulated in MB-Gli1High, suggesting that the deregulation of SHH pathway could be due to the loss of specific miRNAs [56] (Table 7).
![]()
Table 7: MiRNAs profiling studies in medulloblastoma
MB: medulloblastoma, NB: Normal Brain, ANB: Adjacent Normal Brain, NSC: Neural Stem Cells, GNP: Granule Neuron Progenitors, NS: Not Specified, NGS: Next Generation Sequencing, qRT-PCR: quantitative Real-Time Polymerase Chain Reaction. Only miRNAs which showed a fold change greater than or equal to twofold, were considered.
View Table 7
According to miRNAs found consistently deregulated in at least three studies, a putative miRNA profile for MB could be the following: miR-17, miR-21, miR-106b, miR-199b and miR-214 are up-regulated in MB; miR-9, miR-29c, miR-30b, miR-103, miR-124a, miR-128a, mir-129, miR-132, let-7a, let-7e, let-7f are down-regulated in MB.
MiRNA control of Hedgehog signaling in MB
The SHH signaling pathway is a major mitogenic regulator of granule progenitor cells (GPCs) and its deregulation has been implicated in MBs [135]. Figure 3 presented a schematic view of how several functional studied miRNAs could control this oncogenic pathway in MB. Among characterized miRNAs in MB pathogenesis, the over-expression of the miR-17-92 cluster, also known as Oncomir-1, has been deeply investigated. The oncogenic expression of miR-17, miR-19a, miR-20 and miR-92, all of them included in the Oncomir-1 cluster, induces elevated SHH pathway which lead to tumorigenic effects in MB development, among which the c-Myc activation [19,42,135]. Northcott et al. [42] showed a 13q31.3 locus amplification, in which the miR-17/92 cluster mapped. An over-expression of this cluster and its related paralogous (miR-106a/363 and miR-106b/25) was demonstrated in MB if compared to normal human cerebellum [42]. In particular, this over-expression was associated with activated SHH signaling and elevated c-Myc and n-Myc expression levels, supporting the hypothesis that the up-regulation of this miRNA cluster promotes the growth of MB in a SHH-mediated manner [42]. In particular, it is demonstrated that Myc and n-Myc transcriptional regulate miR-17/92 by binding to the promoter of primary miR-17/92 cluster [42,139]. Uziel and colleagues [19] obtained similar results, analyzing miRNAs profiles in rodent models of MB (Ink4c-/-; Ptch +/-; Ink4c-/-; p53-/- genotypes) compared to mature rodent cerebellum and developing cerebellum (P6 GPCs): among the 26 miRNAs that they obtained over-expressed, nine miRNAs belonging to the miR-17-92 cluster. Furthermore, the authors demonstrated that three miRNAs of this cluster, miR-19a, miR-92 and miR-20, were significantly over-expressed in human SHH-active MBs when compared to normal control cerebellum [19].
Ferretti et al. [56] identified a specific signature in tumors with activated SHH (MB-Gli1High): miR-125b, miR-326 and miR-324-5p were down-regulated and the forced over-expression of these three miRNAs inhibited SHH-signaling pathway (Figure 3). In particular, miR-324-5p targets both Smo (the transmembrane protein Smoothened, a SHH-pathway activator) and Gli1 (a downstream transcriptional activator that acts as a SHH-pathway effector) and cooperates with miR-326 to further reinforce the inhibitory activity [56]. The deregulation of these three miRNAs was involved in cell proliferation stimulus through a hedgehog-dependent mechanism [56].
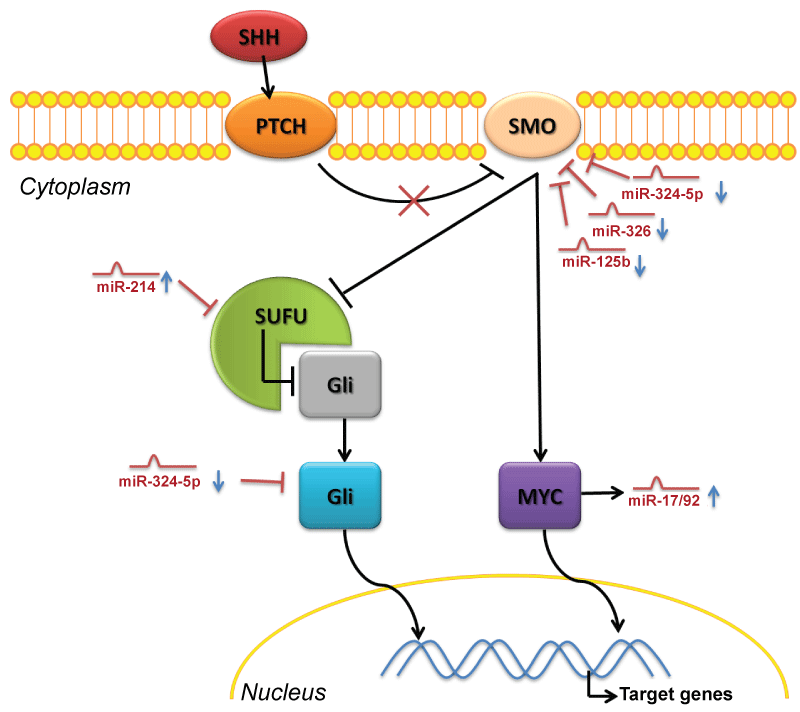
.
Figure 3: MiRNAs control of Sony Hedgehog pathway in medulloblastoma
When SHH binds to its receptor PTCH, it releases its repression on a transmembrane protein Smoothened (Smo) which becomes able to inhibit the repression of SUFU on Gli proteins, which could translocate to the nucleus. A second mechanism induced by activated Smo, by which SHH signaling exerts a proliferative effect, is through the up-regulation of N-Myc. As described in the text, in medulloblastoma the deregulation of several miRNAs support the activate status of this oncogeneic pathway: the lack of miR-324-5p, 326, 125b leads to an over-expression of Smo and Gli factors. In parallel the up-regulation of miR-214 could repress the inhibition of SUFU on Gli proteins. The up-regulation of N-Myc correlates with a deregulation of the Oncomir-1, which is the oncogenic miR-17/92 cluster.
View Figure 3
Finally, miR-214, found up-regulated in human MB by two profiling studies (Table 7) [29,56], could regulate SHH signaling targeting SUFU (Suppressor of fused, a Hh antagonist, which inhibits Gli1 and Gli2 from entering the nucleus) as already demonstrated by Flynt et al. in zebrafish [140].
MiRNA Expression in other Brain Tumors
Tumors of the hematopoietic system
Lymphoma CNS: Primary central nervous system lymphoma (PCNSL) is a rare subtype of extranodal non-Hodgkin's lymphoma that is confined to the CNS in the absence of systemic disease. In patients with PCNSL, definitive diagnosis often is difficult only on the basis of radiographic features and responsiveness to corticosteroids, which both do not specifically distinguish between lymphoma and inflammatory CNS disease [141]. Identification of PCNSL specific miRNAs could provide a potentially useful tool as novel noninvasive biomarker for the diagnosis of this neoplasia (Table 8).
![]()
Table 8: MiRNAs deregulated in "other" brain tumors
DLBCL: Diffuse large B-cell lymphoma; GBM: Glioblastoma, CNS: Central Nervous System, G: Histological Grade, LN: Lymph Node, GH: Growth Hormone, PRL: Prolactin, qRT-PCR: quantitative Real-Time Polymerase Chain Reaction, ISH: In Situ Hybridization.
View Table 8
MiR-21, miR-19b, miR-92a were found up-regulated in a study of 23 patients with PCNSLs and were proposed as marker for PCNSL [21]. These deregulated miRNAs were evaluated also as circulating miRNAs in serum of 14 PCNSL patients. However, no significant differences in the expression levels of miR-21, miR-19b, and miR-92 were observed in serum of PCNSL patients in comparison with that of the control group [21].
MiR-21 and miR-19b were the most abundant miRNAs in 9 CNS diffuse large B-cell lymphomas also in a study by Robertus et al. performed using a qRT-PCR technique [64]. Other than up-regulation of miR-21 and miR-19b, the authors observed also low expression levels of miR-16 and miR-127 [64].
The expression of 13 miRNAs (miR-9, miR-20b, miR-155, miR-340, miR-17-5p, miR-148a, miR-30b, miR-27b, miR-26b, miR-146b, miR-20a, miR-30c and let-7g) was significantly higher in primary CNS lymphomas compared with nodal diffuse large B-cell lymphomas, while five miRNAs (miR-199a, miR-214, miR-432, miR-193b and miR-145) showed a reduced expression in PCNSL if compared with nodal diffuse large B-cell lymphoma. Of the 13 up-regulated miRNAs, the most expressed were miR-9, miR-20b, miR-155, miR-340, miR-17-5p and miR-148a [61]. According to these data, the down-regulation of specific miRNAs, leading to an activation of the MYC (V-Myc Avian Myelocytomatosis Viral Oncogene Homolog) pathway or to a blockade of germinal center exit in B cells, might play a key role in PCNSL pathogenesis [61].
Considering miRNAs found deregulated in at least two papers, diffuse large B-cell lymphomas are characterized by up-regulation of miR-19b and miR-21.
Meningeal tumors
Meningioma: Meningiomas are frequent - accounting for 15% to 20% of all CNS tumors - intracranial neoplasia of adulthood. Most meningiomas are benign (WHO grade I), but certain histological subtypes are associated with less favorable outcomes and correspond to atypical (WHO grade II) and anaplastic (WHO grade III) meningiomas [142].
Five different miRNAs (let-7a, let-7b, let-7c, let-7d and miR-145) were investigated in a series of primary meningioma tissues compared with non-neoplastic arachnoid specimens [36]. A significant down-regulation of miR-145 and let-7d expression in the tumor tissues was observed when compared with non-neoplastic controls. Moreover, lower expression of miR-145 was detected in the group of atypical (WHO grade II) and anaplastic (WHO grade III) meningiomas if compared with benign ones (WHO grade I) [36]. The malignant cell lines transfected with pre-miR-145 were demonstrated to be less invasive and with reduced motility (Table 8) [36].
In a study on 14 human sporadic benign meningiomas compared with arachnoidal tissue controls, 43 miRNAs were found deregulated in most tumor samples. Of the over-expressed miRNAs, miR-335, miR-98, and miR-181a were the most up-regulated (from 25- to 30-fold). Additional miRNAs (miR-19b, let7d, let7g, miR-100, miR-125a, miR-103, miR-370 miR-106b, miR-106a, miR-125b, let7b, miR-26a, miR-23b, and miR-29a) were confirmed to be up-regulated (4- to 10-fold) using qRT-PCR. The three down-regulated miRNAs (miR-200a, miR-373* and miR-575) showed marked decreases [45].
The over-expression of miR-200a in transfected cells showed inhibitory effect on meningioma cell growth, while down-regulation of miR-200a significantly increased, in vitro, the growth of meningioma cells [45].
A TaqMan-based real-time stem-loop RT-PCR analysis in 30 human meningioma specimens compared with normal brain arachnoid tissue, revealed elevated expression levels of miR-335, especially in WHO grade III samples [46]. Moreover, it was demonstrated that miR-335 inhibition lead a statistically significant level of inhibition of cell proliferation compared with that of the control group [46].
The analysis of expression levels of 200 miRNAs from 110 meningioma samples compared to the miRNA expression profiles of 35 normal adjacent tissues revealed that 14 were miRNAs differentially expressed in tumor specimens. Twelve miRNAs (miR-17-5p, miR-22-3p, miR-24-3p, miR-26b- 5p, miR-27a-3p, miR-27b-3p, miR-96-5p, miR-146a-5p, miR-155-5p, miR-186-5p, miR-190a and miR-199a) were up-regulated whereas two miRNAs (miR-29c-3p and miR-219-5p), were significantly down-regulated [54]. Based on their respective miRNA expression profiles, an unsupervised cluster analysis revealed a clear separation of the meningioma samples from the normal adjacent tissues. Moreover, authors observed that miR-96-5p and miR-190a were up-regulated in patients with tumor recurrence if compared to those without recurrence. In contrast, miR-29c-3p and miR-219-5p were up-regulated in patients without recurrence compared to those with tumor recurrence. Finally, the univariate analysis indicated that WHO tumor grade and expression levels of miR-190a, miR-29c-3p and miR-219-5p were significantly correlated with recurrence [54].
Intriguingly, considering all papers about miRNA expression in meningeal tumors, no miRNAs have been found consistently deregulated in at least two papers.
Tumors of the sellar region
Pituitary adenoma: Pituitary adenomas account for at least 10% of intracranial tumors with a prevalence of 1 in 1,000 people in the general population [143].
In a study of eight pituitary adenomas compared with four normal pituitary samples, three miRNAs (miR-124*, miR-515-5p and miR-872) were expressed only in adenoma tissues while five miRNAs (miR-198, miR-299-5p, miR-497*, miR-548c-3p and miR-622) only in normal pituitary tissues. Of the 457 miRNAs expressed in both normal and tumor tissues, 92 were significantly down-regulated and 70 significantly up-regulated in pituitary adenomas [23]. Five miRNAs (miR-135a, miR-140-5p, miR-582-3p, miR-582-5p and miR-938) were predicted to target Smad3 (Small mother against decapentaplegic 3) and all these miRNAs were significantly over-expressed in pituitary adenoma samples. These data support the possibility that the TGF signaling is repressed and modulated by miRNAs in pituitary adenomas via Smad3 whose expression can be directly regulated by several up-regulated miRNAs, including the experimentally validated miR-140 (Table 8) [23].
A microarray assay performed on 12 human human growth hormone-secreting (GH) pituitary adenomas (GH-adenomas) compared normal pituitary gland showed 18 miRNAs down-regulated with at least a 2-fold change in the neoplastic tissues. Conversely, only one miRNA (miR-320) was over-expressed (with a 13.3-fold change) in GH-adenomas [27]. Eight miRNAs (miR-34b, miR-326, miR-374b, miR-432, miR-548c-3p, miR-570 miR-603, and miR-633) were confirmed as down-regulated by qRT-PCR while miR-320 was confirmed as up-regulated [27].
Using bioinformatics tools, three miRNAs (miR-326, miR-432, and miR-570) potentially target the HMGA2 (High Mobility Group AT-Hook 2) gene, while two miRNAs (miR-34b and miR-548c-3p) have both the HMGA1 (High Mobility Group AT-Hook 1) and HMGA2 genes as predicted targets. Finally, miR-326 and miR-603 were predicted to regulate E2F1 (E2F Transcription Factor 1) [27].
A miRNA microarray statistical analysis on 32 pituitary adenomas generated a list of 30 miRNAs differentially expressed between pituitary adenomas and normal pituitary gland [22]. According to expression profile of 29 miRNAs it was possible to predict the different pituitary adenoma histotypes: ACTH- (adrenocorticotropic hormone), GH-, PRL- (prolactin) secreting adenomas and non-functioning adenoma. MiR-26a and miR-149 were found to be up-regulated in pituitary adenomas compared with normal pituitary samples, while miR-21, miR-141 and miR-144 were down-regulated [22].
In an analysis on 21 GH-secreting pituitary adenoma samples (three micro-adenomas and 18 macro-adenomas), 23 miRNAs were found to be up- and 29 down-regulated in GH-adenomas [41]. Moreover, nine miRNAs were differentially expressed between macro- and micro-adenomas: miR-183, miR-193a-5p, miR-222, miR-516b, miR-524-5p, miR-601, miR-629, miR-99b were up-regulated; miR-124, mir-32, miR-574-5p, miR-744 and miR-96 were found to be down-regulated [41].
Trivellin et al. [51] found nine miRNAs (miR-509-3-5p, miR-508-5p, miR-452, miR-330-5p, miR-200a, miR-503, miR-424, miR-449a and miR-199-5p) expressed in normal pituitary gland but not in pituitary adenomas. Different miRNA expressions were observed in different adenoma histotypes. Seven miRNAs (miR-378, miR-516-3p, miR-151-3p, miR-224, miR-618, miR-455-3p and miR-29b) were expressed in all types of adenomas but not in normal pituitary gland. Twelve miRNAs (miR-217, miR-216a, miR-215, miR-502, miR-338, miR-10b, miR-96, miR-202, miR-501, miR-18a, miR-450a and miR-329) were detected only in the non-functioning pituitary adenomas. Five miRNAs (miR-1, miR-760, miR-196b, miR-188-5p and miR-146b-3p) were detected only in the GH-adenoma samples. Three miRNAs (miR-205, miR-132* and miR-523) were detected only in the PRL- adenomas group [51].
Considering miRNAs found deregulated in at least two papers, pituitary adenoma are characterized by up-regulation of miR-516 and down-regulation of miR-548c.
Conclusion
In this review, we have focused on the expression profile of up- or down-regulated miRNAs in brain tumors. MiRNAs have emerged to play a key role in several cellular processes in brain: they are involved in cancer development and progression by regulating gene expression via post-transcriptional regulation, but reliable miRNA profiles of brain tumors are far to be defined. The data reported in the present paper highlight the importance to standardize the tissue source, the analysis methods and the choice of control tissue. In fact, changes in one of these factors could contribute to differences in expression of investigated miRNAs. Identification of tumor-specific miRNAs and of their target genes will improve the knowledge in brain neoplasm and supply additional molecular biomarkers for diagnosis of brain tumors and prospective target therapies. Moreover, the available data provide a basis for in vivo studies to determine the effect of miRNA modulation in chemotherapy. Finally, circulating cancer-related miRNAs could provide an important tool for early non-invasive tumor diagnosis.
References
-
Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, et al. (2006) mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885-1898.
-
Wu L, Fan J, Belasco JG (2006) MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A 103: 4034-4039.
-
Standart N, Jackson RJ (2007) MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes Dev 21: 1975-1982.
-
Lewis BP, CB Burge, DP Bartel (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15-20
-
Ruan K, Fang X, Ouyang G (2009) MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett 285: 116-126.
-
Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857-866.
-
Singh R, Mo YY (2013) Role of microRNAs in breast cancer. Cancer Biol Ther 14: 201-212.
-
Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, et al. (2008) Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A 105: 7004-7009.
-
Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, et al. (2008) Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A 105: 3903-3908.
-
Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, et al. (2007) MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 297: 1901-1908.
-
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, et al. (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114: 97-109.
-
Smith JS, Perry A, Borell TJ, Lee HK, O'Fallon J, et al. (2000) Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol 18: 636-645.
-
Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, et al. (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352: 997-1003.
-
van den Bent MJ, HJ Dubbink, Y Marie, Brandes AA, Taphoorn MJ (2010) IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res 16: 1597-1604.
-
Ciafrè SA, Galardi S, Mangiola A, Ferracin M, Liu CG, et al. (2005) Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 334: 1351-1358.
-
Gal H, Pandi G, Kanner AA, Ram Z, Lithwick-Yanai G, et al. (2008) MIR-451 and Imatinib mesylate inhibit tumor growth of Glioblastoma stem cells. Biochem Biophys Res Commun 376: 86-90.
-
Lang MF, Yang S, Zhao C, Sun G, Murai K, et al. (2012) Genome-wide profiling identified a set of miRNAs that are differentially expressed in glioblastoma stem cells and normal neural stem cells. PLoS One 7: e36248.
-
Loftus JC, Ross JT, Paquette KM, Paulino VM, Nasser S, et al. (2012) miRNA expression profiling in migrating glioblastoma cells: regulation of cell migration and invasion by miR-23b via targeting of Pyk2. PLoS One 7: e39818.
-
Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, et al. (2009) The miR-17~92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci U S A 106: 2812-2817.
-
Zhou X, Ren Y, Moore L, Mei M, You Y, et al. (2010) Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest 90: 144-155.
-
Baraniskin A, Kuhnhenn J, Schlegel U, Chan A, Deckert M, et al. (2011) Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood 117: 3140-3146.
-
Bottoni A, Zatelli MC, Ferracin M, Tagliati F, Piccin D, et al. (2007) Identification of differentially expressed microRNAs by microarray: a possible role for microRNA genes in pituitary adenomas. J Cell Physiol 210: 370-377.
-
Butz H, Likó I, Czirják S, Igaz P, Korbonits M, et al. (2011) MicroRNA profile indicates downregulation of the TGFβ pathway in sporadic non-functioning pituitary adenomas. Pituitary 14: 112-124.
-
Chan JA, Krichevsky AM, Kosik KS (2005) MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65: 6029-6033.
-
Conti A, Aguennouz M, La Torre D, Tomasello C, Cardali S, et al. (2009) miR-21 and 221 upregulation and miR-181b downregulation in human grade II-IV astrocytic tumors. J Neurooncol 93: 325-332.
-
Costa FF, Bischof JM, Vanin EF, Lulla RR, Wang M, et al. (2011) Identification of microRNAs as potential prognostic markers in ependymoma. PLoS One 6: e25114.
-
D'Angelo D, Palmieri D, Mussnich P, Roche M, Wierinckx A, et al. (2012) Altered microRNA expression profile in human pituitary GH adenomas: down-regulation of miRNA targeting HMGA1, HMGA2, and E2F1. J Clin Endocrinol Metab 97: E1128-1138.
-
Dong H, L Luo, S Hong, Siu H, Xiao Y (2010) Integrated analysis of mutations, miRNA and mRNA expression in glioblastoma. BMC Syst Biol 4: 163.
-
Ferretti E, De Smaele E, Po A, Di Marcotullio L, Tosi E, et al. (2009) MicroRNA profiling in human medulloblastoma. Int J Cancer 124: 568-577.
-
Genovesi LA, Carter KW, Gottardo NG, Giles KM, Dallas PB (2011) Integrated analysis of miRNA and mRNA expression in childhood medulloblastoma compared with neural stem cells. PLoS One 6: e23935.
-
Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, et al. (2008) Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res 68: 9125-9130.
-
Guan Y, Mizoguchi M, Yoshimoto K, Hata N, Shono T, et al. (2010) MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin Cancer Res 16: 4289-4297.
-
Hua D, Mo F, Ding D, Li L, Han X, et al. (2012) A catalogue of glioblastoma and brain MicroRNAs identified by deep sequencing. OMICS 16: 690-699.
-
Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, et al. (2009) The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev 23: 1327-1337.
-
Kim G, EC Park, CH Ryu, Sin-Soo Jeon, Seung Il Kim (2011) MicroRNA expression profiling in recurrent anaplastic oligodendroglioma treated with postoperative radiotherapy. JAST 2: 97-104.
-
Kliese N, Gobrecht P, Pachow D, Andrae N, Wilisch-Neumann A, et al. (2013) miRNA-145 is downregulated in atypical and anaplastic meningiomas and negatively regulates motility and proliferation of meningioma cells. Oncogene 32: 4712-4720.
-
Lages E, A Guttin, M El Atifi, Claire Ramus, Hélène Ipas (2011) MicroRNA and target protein patterns reveal physiopathological features of glioma subtypes. PLoS One 6: e20600.
-
Li KK, Yang L, Pang JC, Chan AK, Zhou L, et al. (2013) MIR-137 suppresses growth and invasion, is downregulated in oligodendroglial tumors and targets CSE1L. Brain Pathol 23: 426-439.
-
Liu W, Gong YH, Chao TF, Peng XZ, Yuan JG, et al. (2009) Identification of differentially expressed microRNAs by microarray: a possible role for microRNAs gene in medulloblastomas. Chin Med J (Engl) 122: 2405-2411.
-
Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stühler K, et al. (2010) Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol 20: 539-550.
-
Mao ZG, He DS, Zhou J, Yao B, Xiao WW, et al. (2010) Differential expression of microRNAs in GH-secreting pituitary adenomas. Diagn Pathol 5: 79.
-
Northcott PA, Fernandez-L A, Hagan JP, Ellison DW, Grajkowska W, et al. (2009) The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res 69: 3249-3255.
-
Rao SA, Santosh V, Somasundaram K (2010) Genome-wide expression profiling identifies deregulated miRNAs in malignant astrocytoma. Mod Pathol 23: 1404-1417.
-
Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E (2009) MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer 125: 1407-1413.
-
Saydam O, Shen Y, Würdinger T, Senol O, Boke E, et al. (2009) Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/beta-catenin signaling pathway. Mol Cell Biol 29: 5923-5940.
-
Shi L, Jiang D, Sun G, Wan Y, Zhang S, et al. (2012) miR-335 promotes cell proliferation by directly targeting Rb in meningiomas. J Neurooncol 110: 155-162.
-
Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, et al. (2008) miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med 6: 14.
-
Skalsky RL, Cullen BR (2011) Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS One 6: e24248.
-
Srinivasan S, Patric IR, Somasundaram K (2011) A ten-microRNA expression signature predicts survival in glioblastoma. PLoS One 6: e17438.
-
Teplyuk NM, Mollenhauer B, Gabriely G, Giese A, Kim E, et al. (2012) MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol 14: 689-700.
-
Trivellin G, Butz H, Delhove J, Igreja S, Chahal HS, et al. (2012) MicroRNA miR-107 is overexpressed in pituitary adenomas and inhibits the expression of aryl hydrocarbon receptor-interacting protein in vitro. Am J Physiol Endocrinol Metab 303: E708-719.
-
Yang C, Wang C, Chen X, Chen S, Zhang Y, et al. (2013) Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer 132: 116-127.
-
Zhi F, Chen X, Wang S, Xia X, Shi Y, et al. (2010) The use of hsa-miR-21, hsa-miR-181b and hsa-miR-106a as prognostic indicators of astrocytoma. Eur J Cancer 46: 1640-1649.
-
Zhi F, Zhou G, Wang S, Shi Y, Peng Y, et al. (2013) A microRNA expression signature predicts meningioma recurrence. Int J Cancer 132: 128-136.
-
Zhang W, J Zhang, W Yan, You G, Bao Z (2013) Whole-genome microRNA expression profiling identifies a 5-microRNA signature as a prognostic biomarker in Chinese patients with primary glioblastoma multiforme. Cancer 119: 814-824
-
Ferretti E, De Smaele E, Miele E, Laneve P, Po A, et al. (2008) Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J 27: 2616-2627.
-
Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, et al. (2007) Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA 13: 1668-1674.
-
Weng L, Wu X, Gao H, Mu B, Li X, et al. (2010) MicroRNA profiling of clear cell renal cell carcinoma by whole-genome small RNA deep sequencing of paired frozen and formalin-fixed, paraffin-embedded tissue specimens. J Pathol 222: 41-51.
-
Nonn L, Vaishnav A, Gallagher L, Gann PH (2010) mRNA and micro-RNA expression analysis in laser-capture microdissected prostate biopsies: valuable tool for risk assessment and prevention trials. Exp Mol Pathol 88: 45-51.
-
Hui AB, Shi W, Boutros PC, Miller N, Pintilie M, et al. (2009) Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest 89: 597-606.
-
Fischer L, Hummel M, Korfel A, Lenze D, Joehrens K, et al. (2011) Differential micro-RNA expression in primary CNS and nodal diffuse large B-cell lymphomas. Neuro Oncol 13: 1090-1098.
-
Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, et al. (2006) RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA 12: 187-191.
-
Niyazi M, Zehentmayr F, Niemöller OM, Eigenbrod S, Kretzschmar H, et al. (2011) MiRNA expression patterns predict survival in glioblastoma. Radiat Oncol 6: 153.
-
Robertus JL, Harms G, Blokzijl T, Booman M, de Jong D, et al. (2009) Specific expression of miR-17-5p and miR-127 in testicular and central nervous system diffuse large B-cell lymphoma. Mod Pathol 22: 547-555.
-
Slaby O, Lakomy R, Fadrus P, Hrstka R, Kren L, et al. (2010) MicroRNA-181 family predicts response to concomitant chemoradiotherapy with temozolomide in glioblastoma patients. Neoplasma 57: 264-269.
-
Lakomy R, Sana J, Hankeova S, Fadrus P, Kren L, et al. (2011) MiR-195, miR-196b, miR-181c, miR-21 expression levels and O-6-methylguanine-DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patients. Cancer Sci 102: 2186-2190.
-
de Biase D, Visani M, Morandi L, Marucci G, Taccioli C, et al. (2012) miRNAs expression analysis in paired fresh/frozen and dissected formalin fixed and paraffin embedded glioblastoma using real-time pCR. PLoS One 7: e35596.
-
Visani M, de Biase D, Marucci G, Taccioli C, Baruzzi A, et al. (2013) Definition of miRNAs expression profile in glioblastoma samples: the relevance of non-neoplastic brain reference. PLoS One 8: e55314.
-
van Rooij E (2011) The art of microRNA research. Circ Res 108: 219-234.
-
Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang-Xuan K, et al. (2012) Primary brain tumours in adults. Lancet 379: 1984-1996.
-
Chen Y, Liu W, Chao T, Zhang Y, Yan X, et al. (2008) MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett 272: 197-205.
-
Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, et al. (2008) microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res 68: 3566-3572.
-
Li WB, Ma MW, Dong LJ, Wang F, Chen LX, et al. (2011) MicroRNA-34a targets notch1 and inhibits cell proliferation in glioblastoma multiforme. Cancer Biol Ther 12: 477-483.
-
Smits M, Nilsson J, Mir SE, van der Stoop PM, Hulleman E, et al. (2010) miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget 1: 710-720.
-
Novakova J, Slaby O, Vyzula R, Michalek J (2009) MicroRNA involvement in glioblastoma pathogenesis. Biochem Biophys Res Commun 386: 1-5.
-
Zhang C, Kang C, You Y, Pu P, Yang W, et al. (2009) Co-suppression of miR-221/222 cluster suppresses human glioma cell growth by targeting p27kip1 in vitro and in vivo. Int J Oncol 34: 1653-1660.
-
Tian Y, Nan Y, Han L, Zhang A, Wang G, et al. (2012) MicroRNA miR-451 downregulates the PI3K/AKT pathway through CAB39 in human glioma. Int J Oncol 40: 1105-1112.
-
Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han L, et al. (2010) MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer 9: 229.
-
Pang JC, Kwok WK, Chen Z, Ng HK (2009) Oncogenic role of microRNAs in brain tumors. Acta Neuropathol 117: 599-611.
-
Gillies JK, Lorimer IA (2007) Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle 6: 2005-2009.
-
Lukiw WJ, Cui JG, Li YY, Culicchia F (2009) Up-regulation of micro-RNA-221 (miRNA-221; chr Xp11.3) and caspase-3 accompanies down-regulation of the survivin-1 homolog BIRC1 (NAIP) in glioblastoma multiforme (GBM). J Neurooncol 91: 27-32.
-
Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, et al. (2008) MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res 68: 2773-2780.
-
Quintavalle C, Garofalo M, Zanca C, Romano G, Iaboni M, et al. (2012) miR-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPµ. Oncogene 31: 858-868.
-
Visani M, de Biase D, Marucci G, Cerasoli S, Nigrisoli E, et al. (2014) Expression of 19 microRNAs in glioblastoma and comparison with other brain neoplasia of grades I-III. Mol Oncol 8: 417-430.
-
Karsy M, Arslan E, Moy F (2012) Current Progress on Understanding MicroRNAs in Glioblastoma Multiforme. Genes Cancer 3: 3-15.
-
Lawler S, Chiocca EA (2009) Emerging functions of microRNAs in glioblastoma. J Neurooncol 92: 297-306.
-
Singh SK, Vartanian A, Burrell K, Zadeh G (2012) A microRNA Link to Glioblastoma Heterogeneity. Cancers (Basel) 4: 846-872.
-
Feng SY, Dong CG, Wu WK, Wang XJ, Qiao J, et al. (2012) Lentiviral expression of anti-microRNAs targeting miR-27a inhibits proliferation and invasiveness of U87 glioma cells. Mol Med Rep 6: 275-281.
-
Genovese G, Ergun A, Shukla SA, Campos B, Hanna J, et al. (2012) microRNA regulatory network inference identifies miR-34a as a novel regulator of TGF-ß signaling in glioblastoma. Cancer Discov 2: 736-749.
-
Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, et al. (2009) MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res 69: 7569-7576.
-
Nass D, Rosenwald S, Meiri E, Gilad S, Tabibian-Keissar H, et al. (2009) MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol 19: 375-383.
-
Silber J, Jacobsen A, Ozawa T, Harinath G, Pedraza A, et al. (2012) miR-34a repression in proneural malignant gliomas upregulates expression of its target PDGFRA and promotes tumorigenesis. PLoS One 7: e33844.
-
Tivnan A, Tracey L, Buckley PG, Alcock LC, Davidoff AM, et al. (2011) MicroRNA-34a is a potent tumor suppressor molecule in vivo in neuroblastoma. BMC Cancer 11: 33.
-
Ujifuku K, Mitsutake N, Takakura S, Matsuse M, Saenko V, et al. (2010) miR-195, miR-455-3p and miR-10a( *) are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer Lett 296: 241-248.
-
Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, et al. (2009) Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem 284: 5731-5741.
-
Wong ST, XQ Zhang, JT Zhuang, Chan HL, Li CH (2012) MicroRNA-21 inhibition enhances in vitro chemosensitivity of temozolomide-resistant glioblastoma cells. Anticancer Res 32: 2835-2841.
-
Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, et al. (2008) MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 28: 5369-5380.
-
Gaur AB, Holbeck SL, Colburn NH, Israel MA (2011) Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo. Neuro Oncol 13: 580-590.
-
Lin J, S Teo, DH Lam, Jeyaseelan K, Wang S (2012) MicroRNA-10b pleiotropically regulates invasion, angiogenicity and apoptosis of tumor cells resembling mesenchymal subtype of glioblastoma multiforme. Cell Death Dis 3: e398.
-
Papagiannakopoulos T, Shapiro A, Kosik KS (2008) MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res 68: 8164-8172.
-
Zhang J, Han L, Ge Y, Zhou X, Zhang A, et al. (2010) miR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int J Oncol 36: 913-920.
-
Dews M, Fox JL, Hultine S, Sundaram P, Wang W, et al. (2010) The myc-miR-17~92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res 70: 8233-8246.
-
D'Urso PI, D'Urso OF, Storelli C, Mallardo M, Gianfreda CD, et al. (2012) miR-155 is up-regulated in primary and secondary glioblastoma and promotes tumour growth by inhibiting GABA receptors. Int J Oncol 41: 228-234.
-
Ernst A, Campos B, Meier J, Devens F, Liesenberg F, et al. (2010) De-repression of CTGF via the miR-17-92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene 29: 3411-3422.
-
Fang L, Deng Z, Shatseva T, Yang J, Peng C, et al. (2011) MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-ß8. Oncogene 30: 806-821.
-
Godlewski J, Bronisz A, Nowicki MO, Chiocca EA, Lawler S (2010) microRNA-451: A conditional switch controlling glioma cell proliferation and migration. Cell Cycle 9: 2742-2748.
-
Ma R, Yan W, Zhang G, Lv H, Liu Z, et al. (2012) Upregulation of miR-196b confers a poor prognosis in glioblastoma patients via inducing a proliferative phenotype. PLoS One 7: e38096.
-
Suh SS, Yoo JY, Nuovo GJ, Jeon YJ, Kim S, et al. (2012) MicroRNAs/TP53 feedback circuitry in glioblastoma multiforme. Proc Natl Acad Sci U S A 109: 5316-5321.
-
Tan X, Wang S, Zhu L, Wu C, Yin B, et al. (2012) cAMP response element-binding protein promotes gliomagenesis by modulating the expression of oncogenic microRNA-23a. Proc Natl Acad Sci U S A 109: 15805-15810.
-
Wang K, Wang X, Zou J, Zhang A, Wan Y, et al. (2013) miR-92b controls glioma proliferation and invasion through regulating Wnt/beta-catenin signaling via Nemo-like kinase. Neuro Oncol 15: 578-588.
-
Xia H, Qi Y, Ng SS, Chen X, Chen S, et al. (2009) MicroRNA-15b regulates cell cycle progression by targeting cyclins in glioma cells. Biochem Biophys Res Commun 380: 205-210.
-
Zhang A, Hao J, Wang K, Huang Q, Yu K, et al. (2013) Down-regulation of miR-106b suppresses the growth of human glioma cells. J Neurooncol 112: 179-189.
-
Wang YY, Sun G, Luo H, Wang XF, Lan FM, et al. (2012) MiR-21 modulates hTERT through a STAT3-dependent manner on glioblastoma cell growth. CNS Neurosci Ther 18: 722-728.
-
Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, et al. (2011) Human glioma growth is controlled by microRNA-10b. Cancer Res 71: 3563-3572.
-
Chen G, Zhu W, Shi D, Lv L, Zhang C, et al. (2010) MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol Rep 23: 997-1003.
-
Chen L, Chen XR, Zhang R, Li P, Liu Y, et al. (2013) MicroRNA-107 inhibits glioma cell migration and invasion by modulating Notch2 expression. J Neurooncol 112: 59-66.
-
Chen L, Wang X, Wang H, Li Y, Yan W, et al. (2012) miR-137 is frequently down-regulated in glioblastoma and is a negative regulator of Cox-2. Eur J Cancer 48: 3104-3111.
-
Chen L, Zhang R, Li P, Liu Y, Qin K, et al. (2013) P53-induced microRNA-107 inhibits proliferation of glioma cells and down-regulates the expression of CDK6 and Notch-2. Neurosci Lett 534: 327-332.
-
Deng X, Ma L, Wu M, Zhang G, Jin C, et al. (2013) miR-124 radiosensitizes human glioma cells by targeting CDK4. J Neurooncol 114: 263-274.
-
Fowler A, Thomson D, Giles K, Maleki S, Mreich E, et al. (2011) miR-124a is frequently down-regulated in glioblastoma and is involved in migration and invasion. Eur J Cancer 47: 953-963.
-
Li RY, Chen LC, Zhang HY, Du WZ, Feng Y, et al. (2013) MiR-139 inhibits Mcl-1 expression and potentiates TMZ-induced apoptosis in glioma. CNS Neurosci Ther 19: 477-483.
-
Liu Y, Yan W, Zhang W, Chen L, You G, et al. (2012) MiR-218 reverses high invasiveness of glioblastoma cells by targeting the oncogenic transcription factor LEF1. Oncol Rep 28: 1013-1021.
-
Qu S, Yao Y, Shang C, Xue Y, Ma J, et al. (2012) MicroRNA-330 is an oncogenic factor in glioblastoma cells by regulating SH3GL2 gene. PLoS One 7: e46010.
-
Shi ZM, Wang XF, Qian X, Tao T, Wang L, et al. (2013) MiRNA-181b suppresses IGF-1R and functions as a tumor suppressor gene in gliomas. RNA 19: 552-560.
-
Song L, Huang Q, Chen K, Liu L, Lin C, et al. (2010) miR-218 inhibits the invasive ability of glioma cells by direct downregulation of IKK-ß. Biochem Biophys Res Commun 402: 135-140.
-
Vo DT, Qiao M, Smith AD, Burns SC, Brenner AJ, et al. (2011) The oncogenic RNA-binding protein Musashi1 is regulated by tumor suppressor miRNAs. RNA Biol 8: 817-828.
-
Wu DG, Wang YY, Fan LG, Luo H, Han B, et al. (2011) MicroRNA-7 regulates glioblastoma cell invasion via targeting focal adhesion kinase expression. Chin Med J (Engl) 124: 2616-2621.
-
Wuchty S, Arjona D, Li A, Kotliarov Y, Walling J, et al. (2011) Prediction of Associations between microRNAs and Gene Expression in Glioma Biology. PLoS One 6: e14681.
-
Xia H, Yan Y, Hu M, Wang Y, Wang Y, et al. (2013) MiR-218 sensitizes glioma cells to apoptosis and inhibits tumorigenicity by regulating ECOP-mediated suppression of NF-κB activity. Neuro Oncol 15: 413-422.
-
Hua D, Ding D, Han X, Zhang W, Zhao N, et al. (2012) Human miR-31 targets radixin and inhibits migration and invasion of glioma cells. Oncol Rep 27: 700-706.
-
Cui JG, Zhao Y, Sethi P, Li YY, Mahta A, et al. (2010) Micro-RNA-128 (miRNA-128) down-regulation in glioblastoma targets ARP5 (ANGPTL6), Bmi-1 and E2F-3a, key regulators of brain cell proliferation. J Neurooncol 98: 297-304.
-
Papagiannakopoulos T, Friedmann-Morvinski D, Neveu P, Dugas JC, Gill RM, et al. (2012) Pro-neural miR-128 is a glioma tumor suppressor that targets mitogenic kinases. Oncogene 31: 1884-1895.
-
Shi ZM, Wang J, Yan Z, You YP, Li CY, et al. (2012) MiR-128 inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS One 7: e32709.
-
Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stühler K, et al. (2010) Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol 20: 539-550.
-
Gilbertson RJ (2004) Medulloblastoma: signalling a change in treatment. Lancet Oncol 5: 209-218.
-
Kleihues P, Louis DN, Scheithauer BW, Rorke LB, Reifenberger G, et al. (2002) The WHO classification of tumors of the nervous system. J Neuropathol Exp Neurol 61: 215-225.
-
Brandes AA, Bartolotti M2, Marucci G3, Ghimenton C4, Agati R5, et al. (2015) New perspectives in the treatment of adult medulloblastoma in the era of molecular oncology. Crit Rev Oncol Hematol 94: 348-359.
-
Ajeawung NF, Li B, Kamnasaran D (2010) Translational applications of microRNA genes in medulloblastomas. Clin Invest Med 33: E223-233.
-
O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839-843.
-
Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG (2007) Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet 39: 259-263.
-
Ney DE, Deangelis LM (2010) Management of central nervous system lymphoma, in Non-Hodgkin Lymphomas. Lippincott Williams & Wilkins, Philadelphia: 527-539.
-
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, et al. (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114: 97-109.
-
Daly AF, Burlacu MC, Livadariu E, Beckers A (2007) The epidemiology and management of pituitary incidentalomas. Horm Res 68: 195-198.





