Coronary artery disease (CAD) is a multifactorial disorder. It is important to identify gene mutations that may be responsible for the development of CAD. The aim of this study was to determine the frequency of twelve cardiovascular disease (CVD) related gene mutations in coronary artery patients.
The CVD StripAssay (Vienna Lab, Austria) was performed to analyze the twelve gene mutations on 52 coronary artery patients and 39 healthy controls. After DNA isolation from blood samples, hybridization with biotin marked probes was performed to PCR products containing the related region of mutant genes. Chi-square tests were used for statistical analyses.
Any differences were not observed between coronary artery patients and healthy controls in terms of the frequencies of studied twelve gene mutations except MTHFR A1298C mutation. It was observed that heterozygous and homozygous mutant genotypes of MTHFR A1298C mutation have protective effects against coronary artery disease [p = 0.06, in 95% CI OR = 0.205 (0.0421-1.001)]. Although all the three genotypes of PAI-1 4G/5G, ACE I/D and MTHFR C677T mutations were observed in both groups; homozygous mutant genotypes weren'G20210A, Factor XIII V34L, β-Fibrinojen -455G > A, HPA-1 1a/1b mutations in any of the groups. Heterozygote and homozygote mutant genotypes of APOB R3500Q mutation was not seen in both groups. When the patient group was evaluated in terms of compound genotypes, the rate of two mutations (ACE I/D, MTHFR A1298C) was 56%, the rate of three mutations (PAI-1 4G/5G, MTHFR A1298C, ACE I/D) was 53%, the rate of four mutations (PAI-1 4G/5G, MTHFR A1298C, HPA-1 1a/1b, ACE I/D) was 33%. High cholesterol, the number of clogged arteries and smoking rates was high in CAD patients with compound genotypes or carrying mutations in multiple genes compared the others.
Among 12 studied gene mutations, only the MTHFR A1298C mutation was determined to have a protective effect against CAD. In addition, cumulative effects of the mutant genotypes and environmental factors on people with the combined genotypes may be trigger for CAD.
Gene mutation, Coronary artery, MTHFR, CVD StripAssay
ACE: Angiotensin I Converting Enzyme; APC: Activated Protein C; APOB: Apolipoprotein B; APOE: Apolipoprotein E; CAD: Coronary Artery Diseases; CVD: Cardiovascular Disease; FVL: FV Leiden; HDL: High Density Lipoprotein; HPA1: Human Platelet Antigen 1; LDL: Low Density Lipoprotein; MI: Myocardial Infarction; MTHFR: Methylene Tetrahydrofolate Reductase; PAI-1: Plasminogen Activator Inhibitor 1; PCR: Polymerase Chain Reaction; SNP: Single Nucleotide Polymorphisms; VTE: Venous Thromboembolism
Coronary artery disease (CAD) is a multifactorial, chronic and inflammatory disease in which etiology, environmental and genetic factors play a role [1]. CAD is the leading cause of cardiovascular mortality in the World especially in developing countries [2,3]. Athero-sclerosis in coronary arteries and hypertension have important roles in the development of CAD [4,5]. Hopkins and Williams published a list of over 240 risk factors for CAD [6]. These include smoking, high total cholesterol, increased low density lipoprotein (LDL), decreased high density lipoprotein (HDL), hypertension, stable life, obesity and diabetes. Many genetic risk factors have also been identified [6,7]. Identification of gene variants and their roles that may be responsible for the development of CAD, can play an important role in understanding the key metabolic pathways and pathophysiology of the disease. CAD plays an important role in cardiovascular disease (CVD)'s clinical course [4,8]. For this reason, the CVD related gene mutations that were defined to CVD StripAssay are good candidates for genetic studies of CAD. This strip includes 12 mutations in 10 genes. These genes are Angiotensin I converting enzyme (ACE), apolipoprotein B (APOB), Fibrinogen, Coagulation factor XIII A chain (F13A1, Factor XIII), coagulation factor V (F5, FV), Human Platelet Antigen 1 (HPA1, Gp IIIa, integrin subunit beta 3), Methylene tetrahydrofolate reductase (MTHFR), Plasminogen Activator Inhibitor 1 (PAI-1, SERPINE1), Prothrombin (PTH; F2, coagulation factor II, thrombin) and Apolipoprotein E (APOE).
ACE converts angiotensin I into angiotensin II, an effective vasoconstrictor, which leads to the destruction of vasodilator bradykinin. ACE levels are associated with 287 bp insertion/deletion (I/D) mutation in the ACE gene [9]. I/D mutation indicate a risk factor for myocardial infarction (MI) in older patients and in smokers; the D allele is associated with elevated ACE activity and plasma levels [10].
Familial defective APOB is a cause of autosomal dominant hypercholesterolemia [11]. There is evidence suggesting that various variations of APO B may increase the risk of CAD [12]. R3500Q is a dominant but rare mutation, which is associated with severe hypercholesterolemia and elevated risk for atherosclerosis [11].
Fibrinogen is a glycoprotein located in the long arm of chromosome 4 (4q23-32) and containing 2 copies of 3 polypeptide chains encoded by 3 different genes (α, β, γ) [13]. Several studies have shown that variations of the fibrinogen beta chain (FGB, β-fibrinogen) gene are associated with increased severity of CAD [14,15]. -455G > A mutation increases the risk for premature MI and ischemic stroke [16]. In most studies it was shown that those carrying the -455A allele have higher plasma fibrinogen levels [14]. This leads to a large clot formation and an increased risk of arterial thrombosis.
Factor XIII is a transglutaminase. Val34Leu (V34L) mutation in the F13A1 gene occurs at the cleavage site of the F13A1 activation peptide. The mutation causes enzyme deficiency and bleeding disorders [17,18]. The L variant offers a protective effect against venous thromboembolism (VTE) [19].
Single nucleotide polymorphisms (SNP) in the F5 gene have been shown to play a major role in CAD development [20]. FV Leiden (FVL) (1691G > A; R506Q) mutation in F5 destroys the major proteolytic cleavage site recognized by activated protein C (APC), and FV becomes resistant to APC, but has normal procoagulant activity [21]. FVL inactivates 10 times slower than normal F5 and remains in circulation longer and a predisposition to coagulation occurs [21]. The risk of VTE, peripheral vascular diseases, stroke, pulmonary embolism increases in individuals with FVL mutation and it also causes recurrent miscarriages, second and third trimester pregnancy losses, preeclampsia, placental abrasion, intrauterine growth retardation [22]. FV R2 haplotype (H1299R) have a mild risk factor for thrombosis; increases CVD risk for carriers of FV Leiden [23].
HPA1 gene 1a/1b (L33P) mutation has been associated with acute coronary events, MI and stroke predisposition at early ages [24].
MTHFR is an important enzyme in folate metabolism. The C677T and A1298C mutations of MTHFR gene have been described as risk factors for CAD, stroke, neural tube defects, Down syndrome, breast and endometrial cancers [25].
In the promoter region (675 base pairs prior to the start of transcription) of the PAI-1 gene, an insertion or deletion mutation (4G/5G) was detected [26]. The 4G allele is associated with higher PAI-1 transcription rates [26]. An increase in plasma PAI-1 level could damage the fibrinolytic system and lead to a permanent fibrin clot. Therefore, this mutation may help to evaluate the performance of the fibrinolytic system in the diagnosis of CAD.
The G20210A mutation in the 3'-untranslated region of the PTH gene was reported to be associated with increased prothrombin levels in plasma and a tendency to thrombosis [27].
The affinity of lipoproteins containing APOE to lipoprotein receptors increases from APOE-E2 to APOE-E3 and APOE-E4. As the allele number increases (E2, E3 and E4) APOE decreases and plasma cholesterol, LDL-cholesterol and APO B increase [28].
In our study, we aimed to investigate the mutations of Factor V, Prothrombin, Factor XIII, β-Fibrinogen, PAI-1, HPA1, MTHFR, ACE, APOB and APOE genes in patients with coronary artery disease.
In this study, a total of 92 individuals, 52 coronary artery patients (47 males and 5 females) aged between 32 and 54 years (mean age ± SD: 47.6 ± 5.8 years) and 39 healthy controls (24 females and 15 males) aged between 19 and 60 years (mean age ± SD: 44.8 ± 9.8 years) were examined. The CAD patients were treated and followed up in the Cardiology Department of Ondokuz Mayis University Research Hospital, Samsun, Turkey. All the unrelated participants (patients and controls) were from Northern Turkey. The clinical data of patients including hypertension, diabetes, hypercholesterolemia, family history of CAD, smoking habits and number of diseased vessels were recorded (Table 1). Coronary risk factors were defined according to following criterions: hypertension was defined if the patients' blood pressure was > 140/90 mmHg or if they were receiving any antihypertensive treatment; diabetes was defined if they were receiving any antidiabetic medication; hypercholesterolemia, was considered when their triglyceride concentration was > 200 mg/dL, their total cholesterol concentration was > 200 mg/dL, or they were receiving lipid-lowering drugs. Smoking habit was defined as never and current smoker. The family history of CAD was defined as positive if at least one first-degree relative was diagnosed with CAD. Coronary angiography was applied to all patients. If the luminal diameter of at least one epicardial coronary artery was narrowed by > 50%, coronary stenosis was accepted significant. The exclusion critters for control group were hypertension, diabetes, hypercholesterolemia and family history of CAD and CVD.
Table 1: Clinical characteristics of CAD patients. View Table 1
The study was approved by the Ondokuz Mayıs University Clinical Research Ethics Committee with the decision number 2011/330 and informed consent was obtained from the patient and control group volunteers.
DNA was purified by salting out method from 5 ml blood samples and stored at -20 ℃. Polymerase chain reaction (PCR) and reverse-hybridization kit (Vienna Lab CVD Strip Assay Kit) assays done for mutation analysis according to the instructions described by the manufacturer (ViennaLab Labordiagnostika GmbH, Vienna, Austria, www.viennalab.com) Shortly, for amplifying different gene sequences, a multiplex PCR in a single reaction tube were made and simultaneously biotin-labeled. The PCR conditions were as follows: 94 ℃ for 2 min, followed by 35 cycles of 94 ℃ for 15 s, 58 ℃ for 30 s, 72 ℃ for 30 s, and a final extension step of 72 ℃ for 3 min. Lastly, the PCR products were selectively hybridized to a test strip that include allele-specific oligonucleotide probes immobilized as an array of parallel lines. Then streptavidin-alkaline phosphatase and color substrates were used to detect bound biotinylated sequences. The CVD strip, demonstrating the band patterns of the mutant type and wild type alleles of the genes, is shown in Figure 1. For the genotypes of normal individuals, only wild bands should be positive, for heterozygous individuals both wild and mutant bands, and for homozygous mutant individuals only mutant bands should be positive. Six possible homozygous and heterozygous APOE genotypes (E2/2, E3/3, E4/4, E2/3, E2/4, and E3/4) could be obtained for the APOE isoforms E2, E3 and E4.
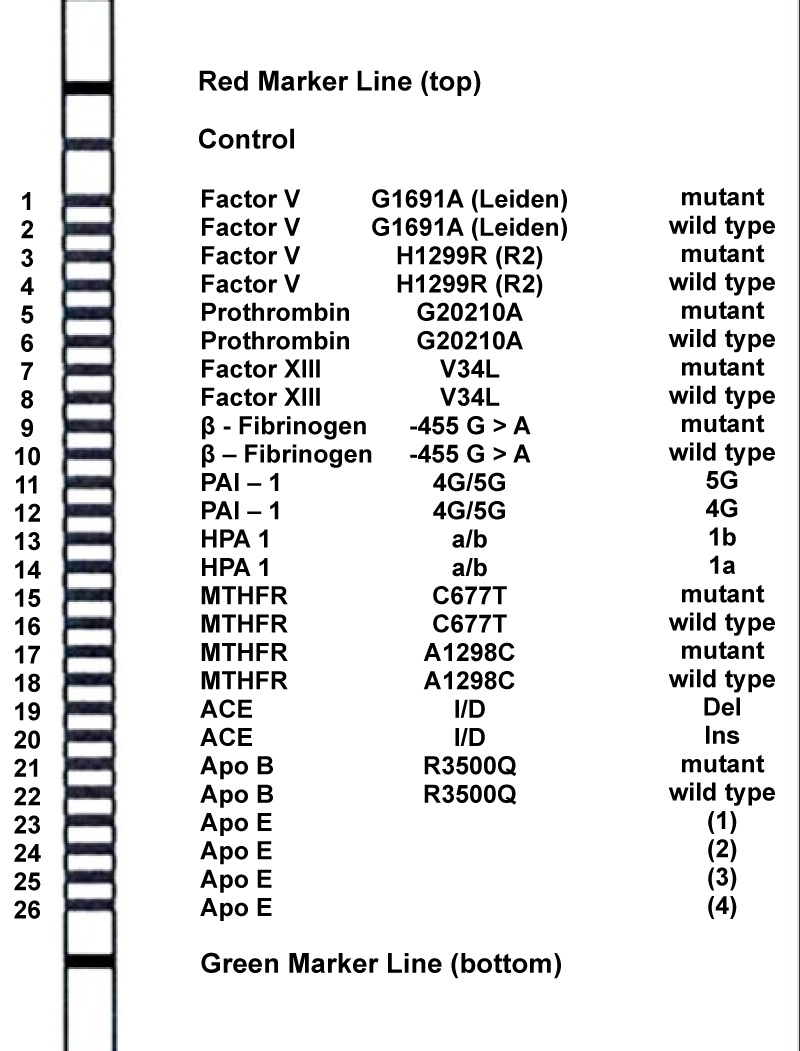 Figure 1: Band paterns of 26 bands in CVD Strip (Vienna Lab).
View Figure 1
Figure 1: Band paterns of 26 bands in CVD Strip (Vienna Lab).
View Figure 1
As a result of the analysis of the patients and controls, mutation distributions in the related genes were estimated by using SPSS 15.0 statistical program. Gene mutations and compound genotypes in coronary artery patients and healthy controls were compared by chi-square analysis (Open Epi Version 3.03).
CVD StripAssay results and genotypes for twelve mutations of two cases are shown in Figure 2. The frequency of related gene mutations for all coronary artery patients and healthy controls are shown in Table 2. According to the results, except MTHFR A1298C mutation, any differences were not observed between coronary artery patients and healthy controls in terms of the frequencies of studied gene mutations. Coronary artery patients had homozygous wild-type genotype for MTHFR A1298C significantly more often than healthy controls (p = 0.008). When AA: AC + CC genotypes were compared in the patient and control groups, heterozygous and homozygous mutant genotypes had shown to have protective effects against CAD [p = 0.06, OR = 0.205 (0.0421-1.001) in 95% confidence interval]. Homozygous mutant genotypes weren'G20210A, Factor XIII V34L, β-Fibrinojen -455G > A, HPA-1 1a/1b mutations in both groups. All the three genotypes of PAI-1 4G/5G, ACE I/D and MTHFR C677T mutations were observed in cases and controls. Heterozygote and homozygote mutant genotypes of APOB R3500Q mutation was not seen in any of the groups.
 Figure 2: CVD StripAssay results of two cases. Both cases are homozygote wild type for Factor V Leiden, Prothrombin G20210A, Factor XIII V34L, MTHFR C677T and APOB R3500Q; Number 1 is heterozygote and Number 2 is homozygote wild type for Factor V H1299R and MTHFR A1298C; Number 1 is homozygote wild type and Number 2 is heterozygote for β-Fibrinojen -455G > A; Number 1 is 5G/5G and number 2 is 4G/4G for PAI-1; both the cases are 1a/1a for HPA-1; Number 1 is del/del and Number 2 is del/ins for ACE; both the cases are E3/E3 for APOE.
View Figure 2
Figure 2: CVD StripAssay results of two cases. Both cases are homozygote wild type for Factor V Leiden, Prothrombin G20210A, Factor XIII V34L, MTHFR C677T and APOB R3500Q; Number 1 is heterozygote and Number 2 is homozygote wild type for Factor V H1299R and MTHFR A1298C; Number 1 is homozygote wild type and Number 2 is heterozygote for β-Fibrinojen -455G > A; Number 1 is 5G/5G and number 2 is 4G/4G for PAI-1; both the cases are 1a/1a for HPA-1; Number 1 is del/del and Number 2 is del/ins for ACE; both the cases are E3/E3 for APOE.
View Figure 2
Table 2: The genotypic frequencies of twelve mutations in ten cardiovascular disease genes in coronary artery patients and healthy control groups. View Table 2
When the mutations observed in the patient group were evaluated, it was observed that there were individuals carrying more than one mutation in different genes. These include; in 26 (56%) of 46 patients, two mutations (ACE I/D, MTHFR A1298C), in 8 (53%) of 15 patients, three mutations (PAI-1 4G/5G, MTHFR A1298C, ACE I/D) and in 5 (33%) of 15 patients four mutations (PAI-1 4G/5G, MTHFR A1298C, HPA-1 1a/1b, ACE I/D) were found to be transported together (Table 3).
Table 3: Patients with Compound Genotype. View Table 3
CAD is a multifactorial disease and multifactorial diseases are caused by the interaction of genetic and environmental factors. Many genetic factors that cause coronary artery disease have been identified [7]. CAD plays an important role in clinical course of CVD [4,8]. For this reason, the CVD related gene mutations that were defined to CVD StripAssay are good candidates for genetic studies of CAD. In the current study, twelve gene mutations in the Factor V, Prothrombin, Factor XIII, β-Fibrinogen, PAI-1, HPA1, MTHFR, ACE, APOB and APOE genes were analyzed by CVD StripAssay on study population.
The frequency of FVL mutation in the normal population is generally 2-4%. However, it was reported to be 3% in Europe, 4.4% in America, 14% in Greece, 7.1-10.3% in Turkey, and 12.2% in Northern Cyprus [23,29,30]. In our study, homozygote wild type and heterozygote frequencies were found to be 96.3% and 3.7% in the coronary artery patients group, respectively. In the control group, only homozygote wild type genotype was observed. Homozygote mutant genotype was not found in both groups. In another study from Turkish population including 4,709 individuals with cardiovascular disease, the frequency of FVL mutation was observed 0.57% in homozygous form and 7.43% in heterozygous form [31]. Close to the rates observed in this study, homozygous mutant genotype was not observed in our study. Although the number of samples in which this gene was studied was limited, the presence of mutant alleles in CAD patients, but not in healthy controls, revealed a possible relationship between CAD and FVL mutation.
In different regions of the world, the frequency of Arg allele was reported to be 9.5-16.5% in VTE cases; 5.8-10.4% in healthy controls [32-37]. In our study, we didn't observed any Arg/Arg genotype in the analysis of His1299Arg mutation in both groups. In our study, the His/His and His/Arg genotype frequencies were 83.3% and 16.7% in the coronary artery patients group and 85.7% and 14.3% in the healthy control group, respectively. Although in our study the sample size was limited, the rate of Arg allele in the patient group (8.3%) was smaller than the findings of worldwide, but the frequency of Arg allele in our control group (7.1%) was found to be similar to the findings of worldwide. In another study with the similar number of samples we studied, in order to investigate the role of various homeostatic gene polymorphisms in young patients in Egypt, including 31 MI, 21 unstable angina and 20 controls, the frequency of H1299R mutation was found to have no difference between patients and controls similar to our results [38].
In our study, only GG and GA genotypes were observed among the genotypes identified as GG, GA and AA in the prothrombin gene. While 29 (96.7%) of 30 patients with coronary artery disease had wild type GG, 1 (3.3%) had heterozygous GA genotype, in the control group which was very low in size, 6 of 7 samples had GG and 1 had GA genotype. No AA genotype was found in either group. Any statistically significant difference was not observed between patients and controls in terms of Prothrombin G20210A mutation. In another study including 96 MI patients and 77 controls from the south region of Turkey, Dönmez, et al. has been reported that there was no relationship between MI development and this variant (P: 0.4) [39]. According to results of another study from Turkey including 4,709 patients, the frequency of mutation in the homozygous form was found 0.25%, and in heterozygous form it was found to be 3.44% similar to our results [31].
The frequency of Factor XIII Val34Leu mutation is 25% in the white, lower in black people and Asian Indians, and extremely rare in the Japanese population [40]. In a separate case-control study in Hungary, FXIII-Leu34 amino acid exchange was found to provide protection against MI [40,41]. A study in the Aegean Region in Turkey supports the FXIII Val/Leu genotype has a protective effect from MI [42]. In our study, among the genotypes identified as Val/Val, Val/Leu and Leu/Leu in the Factor XIII gene, 6 of 7 patients had homozygous Val genotype, 1 patient had heterozygous Val/Leu genotype and 4 individuals in the control group had homozygous Val genotype. Homozygous Leu genotype was not found in both groups. Unlike other mutations, there was no evidence for the protective effect of the Val/Leu genotype due to the limited number of samples studied for the Val34Leu mutation. In our study for factor XIII mutation, the loss data due to the Strip Assay were quite high.
In our study, only GG and GA genotypes were observed in the genotypes identified as GG, GA and AA in the β-fibrinogen gene and their frequency was 66.7% and 33.3% in the coronary artery patient group, and 83.3% and 16.7% in the control group, respectively. No AA homozygous mutant genotype was observed in either group. No association was found between AA genotype and CAD in our study. In a study conducted in Kayseri in Turkey, which included 35 coronary artery patients, -455A alleles were found to have no effect on the development and severity of CAD [43]. In a similar study in Poland, 426 ischemic stroke patients and 234 controls were examined. Genotype frequencies were GG 75%, GA 36.8%, AA 6.6% in patient group and GG 5.3%, GA 32.9% and AA 9.8% in control group, and no significant difference was found between the patient and control groups in terms of this mutation [44]. GA genotype frequency, which is 33.3% in our patient group, is similar to the genotype frequency in the study conducted in Poland.
In our study, the frequency of genotypes identified as 5G/5G, 4G/4G and 5G/4G were 42.1%, 15.8%, 42.1% in the coronary artery patient group and 57.1%, 2.6%, 14.3% in the control group respectively. It was observed that 4G allele was higher in the coronary artery group. Similar to our findings, in a study including 127 MI and 127 controls in Mexico, it was found that 4G allele was an independent risk factor for young acute MI in smokers, hypertension patients and that having family history [45]. In another study involving 305 MI patients and 328 controls, the risk of MI was found to be high with increased plasma PAI-1 levels in 4G carriers [46].
In our study, only 1a/1a and 1a/1b genotypes of the genotypes identified as 1a/1a, 1a/1b and 1b/1b in the HPA1 gene were observed. These genotype frequencies were 66.7%, 33.3% in coronary artery patient group, respectively. In the control group, only the 1a/1a genotype was observed. Any 1b/1b genotype was found in either group. Some studies have shown that HPA-1b homozygote individuals have high risk for ischemic cardiovascular disease and MI [47,48]. In contrast, a study from Sweden including 369 patients and 388 controls revealed that HPA-1 gene mutation was not associated with coronary atherosclerosis or MI [49]. In addition, studies including 510 CAD patients in the USA and 1211 CAD patients in Germany have reported that the HPA-1b allele causes early MI but is not associated with CAD [50]. These differences may be due to factors such as the number of samples studied or ethnic origin.
In our study, the frequency of genotypes identified as CC, CT and TT for MTHFR C677T mutation were estimated as 60%, 20% and 20% in coronary artery patients group, 57.1%, 28.6% and 14.3% in control group, respectively. Although the frequency of TT genotype was higher in the patient group compared to the control group, a complete association with CAD could not be made due to the limited number of samples. In another study from Turkey conducted by Yılmaz, et al. with 93 healthy controls and 79 coronary artery patients for 677C > T mutation, no statistically significant difference was found between patient and control groups in terms of this mutation (p > 0.05) [51]. In contrast to this study, another study conducted in China, involving 694 hyperlipidemia patients and 897 controls, found that those carrying the TT genotype had higher plasma homocysteine values (p < 0.01) [52].
In our study regarding the MTHFR A1298C mutation, the frequency of genotypes identified as AA, AC, CC were 20.8%, 77.1%, 2.1% in the coronary artery patient group, 5.1%, 94.9%, 0% in the control group, respectively. When AA: AC + CC genotypes were compared between patient and control groups, heterozygous and homozygous mutant genotypes had shown to have protective effects against CAD [p = 0.06, OR = 0.205 (0.0421-1.001) in 95% confidence interval]. In another study conducted in Turkey including 86 coronary artery patients and 90 controls, no correlation was found between CAD and A1298C mutation [53]. However, in another study performed in Turkey with 79 conotruncal heart patients and 99 controls, AC and CC genotypes were observed to be significantly higher in patient group and A1298C mutation was found to be a risk factor for conotruncal heart disease (p < 0.05) [54].
In our study, the frequency of genotypes identified as II, DI and DD in the ACE gene were determined as 4.7%, 69.8%, 25.6% in coronary artery patients group, and 8%, 68%, 24% in the control group, respectively. DI and DD genotype frequencies were similar in both groups. When the genotype frequencies of both groups were compared, no correlation was found between this mutation and CAD [p = 0.89, OR = 1.089 (0.346-3.42)]. In a study conducted in Iran including 676 patients with coronary artery disease and 374 controls, ACE I/D mutation had found to have no effect on the development of CAD, similar to our findings [55]. However, in a study conducted in Turkey with 203 coronary artery patients and 140 controls, DD genotype was found to be higher in the patient group [56].
In our study, heterozygote and homozygote mutant genotypes of APOB R3500Q mutation was not observed in both groups. In a similar study involving 163 CAD patients and 114 controls held in Turkey, APOB R3500Q mutation was not observed in both groups, either [57]. In another study from Lebanon, this mutation has not been observed and it has been reported that this mutation may increasingly be lost in Mediterranean countries such as Israel, Spain and Turkey [58].
In our study, the frequency of E2/E3 genotype was 11.1% in the coronary artery patients group and 0% in the control group; the frequency of E3/E3 genotype was 83.3% in the coronary artery patients group and 90.9% in the control group; the frequency of E3/E4 genotype was 5.6% in the coronary artery patients group and 9.1% in the control group. Other genotypes were not found in either groups. The relationship with CAD and APOE mutation was not significant because there was no numerical match between patient-control groups due to data loss. In contrast to our study, E4 allele was found to be higher in another study from Turkey including 70 children with coronary artery disease and 67 controls and also in children with E4 allele, T-Arm and LDL levels were found to be high [59]. In a meta-analysis including 6634 CAD patients and 6394 controls from China, it was reported that the APOE E4 allele may be a risk factor for CAD [60].
When the mutations observed in the patient group were evaluated, it was observed that there were individuals carrying more than one mutation. These include; 26 of the 46 patients (56%) had two mutations (ACE I/D, MTHFR A1298C); 8 of 15 patients (53%) have three mutations (PAI-1 4G/5G, MTHFR A1298C, ACE I/D), 5 of 15 patients (33%) have four mutations (PAI-1 4G/5G, MTHFR A1298C, HPA-1 1a/1b, ACE I/D). These findings suggest that the mutation of more than one gene in a person is a trigger for CAD due to the cumulative effect. It was found that high cholesterol, obstructed vessel count and smoking were higher in patients with CAD who had compound genotype or mutations in more than one gene.
One of the most important limitations of the current study is the small sample size and this lowers the statistical power. Because of the methodology and financial restrictions, we studied limited number of cases.
In conclusion, among the twelve mutations studied only the A1298C mutation in MTHFR gene showed an association with CAD. It was observed that the frequencies of heterozygous and homozygous mutant genotypes of A1298C mutation in MTHFR gene were significantly higher in control group than CAD patients. Based on this result, we can say that A1298C mutation in MTHFR gene had a protective effect against CAD. Furthermore, it was indicated that the cumulative effect of mutant genotypes and environmental factors may be a trigger for CAD in individuals with compound genotypes. To our knowledge it is the first study that investigated 12 mutations in ten cardiovascular related genes in CAD patients. In the mutation analysis of the 10 genes studied, we think that if the number of patients and controls is expanded, more predictive results can be obtained for CAD.
This study was supported by the Ondokuz Mayıs Uni¬versity Research Fund (Project No: PYO.TIP.1904.12.001) and is a report of Master of Science thesis.
The authors declare that they have no conflict of interest.