Objectives: This study aimed to investigate the diagnostic value of peripheral microRNA (miRNA) expression in schizophrenia (SZ).
Methods: By using an Affymetrix array to identify differentially expressed miRNAs in SZ patients; quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) was used to verify identified microRNA and test major depressive disorder (MDD), generalized anxiety disorder (GAD) and mental retardation (MD) related microRNAs for comparison.
Results: The expression levels of miR-1972, miR-26b, miR-4485, miR-4498 and miR-4743 were up-regulated significantly in MDD patients, among which the expression levels of miR-26b, miR-4485 and miR-4743 were also up-regulated significantly in GAD patients, and miR-4485 and miR-4743 upregulated in MD patients; The ROC curve of miR-26b in SZ patients showed that its sensitivity and specificity for diagnosis were 0.721 and 0.950 respectively with the area under curve (AUC) being 0.868; the ROC of miR-26b for SZ and MD differentiation showed that its sensitivity and specificity were 0.78 and 0.65 respectively with AUC being 0.765; the ROC of miR-26b for SZ and GAD differentiation showed that its sensitivity and specificity were 0.667 and 0.825 respectively with AUC being 0.802; the ROC of miR-26b for SZ and MDD differentiation showed that its sensitivity and specificity were 0.537 and 0.675 respectively with AUC being 0.629.
Conclusions: MiR-26b might have significant diagnostic value for SZ, and probably also serve a significant role in MDD.
MicroRNA, SZ, MDD, MD, GAD, Diagnostic value
Schizophrenia (SZ) is a common and often disabling mental illness characterized not only by a varied group of clinical symptoms, but also wide-ranging deficits in neurocognitive and neurophysiological functions [1]. Recent genome-wide association studies (GWASs) of SZ have identified more than 100 risk loci [2]. One study found that the well-known schizophrenia-associated deletion at 22q11.2 includes both miR-185 as well as the gene DGCR8, a component of the microprocessor complex that is involved in the initial step of miRNA biogenesis [3]. The loss-of-function mutation of AGO2 results in global developmental defects, with the most prominent malformation found in the nervous system, this suggested that Ago2-dependent pre-miRNA processing is particularly important for the biogenesis of miRNA in this cluster and may be associated with changes we observed in schizophrenia [4,5]. Though numerous genes have been implicated in SZ susceptibility, with the pathophysiology of schizophrenia (SZ) being unknown and genetic heterogeneity being a fact, the search for SZ risk genes has proven very difficult. The scarcity of tangible pathology determines that clinical diagnosis could only be based on descriptive criteria and requires persistent or episodic presence of certain symptoms over a defined period with associated disability and exclusion of other psychiatric conditions [6]. As is known, early intervention in schizophrenia is thought to be important in improving outcome, any delay in diagnosis or misdiagnosis owing to the inherent limitations of clinical descriptors may have a negative prognostic impact [7]. Therefore, accessible biological markers are urgently needed to improve the timing and accuracy of diagnosis, and identify individuals at very high risk of schizophrenia.
MicroRNAs (miRNAs), small RNAs approximately 19-23 nucleotides long, with complementarity to specific regions in messenger RNAs (mRNAs), are key regulators of gene expression via binding of miRNAs to target mRNAs triggers the cleavage, translational repression, or deadenylation of these targets, miRNAs regulate gene expression at the post-transcriptional level and are important for coordinating nervous system development and neuronal function in the mature brain [8,9]. It has been increasingly accepted that a link between altered miRNAs and risk of neuropsychiatric disorders has been established. Recent work and our previous studies have revealed that abnormal levels of miRNAs are present in the brains of patients with neuropsychiatric disorders, such as schizophrenia [10], major depressive disorder (MDD) [11], mental retardation (MD) [12] and generalized anxiety disorder (GAD) [13]. Not many studies on association between miRNA expression and GAD or mental MD were found until now. It was generally argued that environmental and genetic factors all contribute to its development, genetic factor in particular, we can say that miRNAs are likely to be pleiotropic and may contribute to susceptibility to neuropsychiatric disorders.
In this study, we selected 5 miRNAs identified in MDD for testing in SZ, GAD and MD patients, in an effort to identify certain miRNA(s) as biomarker for psychotic diseases.
All individuals recruited in the study were provided with written informed consent. The study was approved by local Institutional Review Boards. Between Aug. 2018 and Jun. 2020, 40 SZ patients (19 male and 21 female), aged from 15 to 63 with an average age of 30.98 + 12.96, 81 MDD patients (33 male and 48 female), aged from 15 to 68 with an average age of 33.62 ± 15.17, 30 GAD patients (7 male and 23 female), aged from 11 to 80 with an average age of 46.60 ± 14.55, 25 male MD patients, aged from 9 to 26 with an average age of 16.44 ± 4.46, fulfilling the criteria as defined by the Diagnostic and Statistical Manual 5th edition (DSM-V), were prospectively recruited from No. 904 Hospital of PLA. Clinical diagnoses of the patients were made by at least two consultant psychiatrists, and the diagnoses were further confirmed by an additional experienced clinical psychiatrist. All SZ, MDD and GAD patients were first visitors to the clinics and prior to any clinical treatment, or in the absence of antipsychotics within at least three months. No patient had history of severe medical diseases, other psychiatric disorders, structural brain disorders, mental retardation, unstable psychiatric features and movement disorders. Also, patients who had brain injury causing traumatic amnesia longer than 24 hours and who received blood transfusion within a month or electroconvulsive therapy within 6 months, were excluded from the study. All MD patients had IQ score below 70, less than 8 for those below 16-years-old by Children Adaptive Capacity Scale, and more than 7 for those above 16 years old by Adult Intellectually Disability Assessment Scale. All the following medical conditions are ruled out for MD patients, including perinatal brain injury, perinatal cerebral anoxia, ischemia, infection and trauma, karyotype abnormalities, Genetic metabolic diseases, Hereditary neurodegenerative disorders, fragile X syndrome, maternal alcoholism and drug addiction during pregnancy, and any other psychiatric or medical disorders.
In addition, 43 healthy controls for SZ (20 male and 23 female), aged from 16 to 64 with an average age of 29.40 + 12.49, 46 healthy controls for MDD (20 male and 26 female), aged from 15 to 68 with an average age of 32.59 ± 14.79, and 30 healthy controls for GAD (8 male and 22 female), aged from 21 to 71 with an average age of 46.43 ± 13.03, without any family history of other major psychiatric disorders within the last three generations were recruited. Similarly, all healthy controls were without any history of blood transfusion or severe traumatic event within a month. Another 25 male healthy controls for MD, aged from 7 to 26 (20 ± 6), were also recruited.
Whole blood (5 ml) was collected from each subject using EDTA anticoagulant tube and processed within 3 hours. The plasma was separated by centrifugation, then transferred into fresh RNase/DNase-free 2 ml microcentrifuge tube, and stored at -80 °C until use. Total RNAs were isolated from the plasma with the mirVana™ PARIS™ Kit (Applied Biosystems, p/nAM1556). The RNA concentration and quality was measured with a NanoDrop ND-2100 spectrophotometer (Thermo Scientific). The RNA integrity was assessed using Agilent 2100 (Agilent Technologies). To ensure a robust analysis for the following procedures, samples with an RNA integrity number (RIN) inferior to 8 were excluded.
RNA samples from three SZ, MDD, GAD and MD patients and three corresponding controls were used for miRNA microarray profiling. MiRNA expression was measured by Affymetrix miRNA 3.0 array (Affymetrix, Santa Clara, CA, USA) containing probes for a total of 723 human miRNAs. The sample labeling, microarray hybridization and washing were performed based on the manufacturer's standard protocols. Briefly, total RNA were tailed with Poly A and then labeled with Biotin. Afterwards, the labeled RNAs were hybridized onto the microarray. Having washed and stained the slides, the arrays were scanned by the Affymetrix Scanner 3000 (Affymetrix). The scanned images were analyzed using Expression Console software (version1.3.1, Affymetrix).
According to microarray results, the significantly differentially expressed miRNAs from SZ, MDD, GAD and MD patients were chosen for further validation with real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR). Total RNAs were extracted from the purified plasma using mirVana™ PARIS™ Kit (Applied Biosystems, p/nAM1556) for quantitative detection of miRNA. Complementary DNA was synthesized using the Reverse Transcription TaqMan MicroRNA Reverse Transcription Kit and miRNA-specific stemloop primers (Applied Biosystems, inc., USA, P/N: 4366596) according to the manufacturer's instructions. The RT reaction consisted of 5 μL of total RNA (~10 ng), 0.15 μL dNTPs with dTTP (100 mM), 1.00 μL multiscribe RT enzyme (50 U/ul), 1.5 μL 10 × RT Buffer, 0.19 μL RNase Inhibitor (20 U/μL), 4.16 μL nuclease-free water, 3 μL 5 × RT primer, in a total volume of 15 μL. Reactions were performed using the following conditions: 16 °C for 30 min, 42 °C for 30 min, 85 °C for 5 min, and held at 4 °C. Real-time PCR reactions were performed using Applied Biosystems 7900HT Real-Time PCR System (Applied Biosystems, Inc., USA), with 10 μL PCR reaction mixture that included 2 μL of the cDNA, 5 μL of 2 × TaqMan Universal PCR Master Mix II (Applied Biosystems, Inc.), 0.5 μL of 20 × miRNA-specific PCR primer/probe mix (Applied Biosystems, inc.) and 2.5 μL of nuclease-free water. PCR reactions in a 384-well plate were run at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 1 min. Each sample was run in triplicate for analysis. The 5 × RT primers (miRNA-specific stem-loop primers) and 20 × miRNA-specific PCR primer/probe mix were supplied by the TaqMan MicroRNA Assays (Applied Biosystems, Inc.) based on the miRNA sequences obtained from the miRBase database. Data were collected using the SDS 2.3 software (Applied Biosystems, Inc.). After normalized to RNU48, the expression levels of miRNAs were calculated using the 2-ΔΔCt method.
At least 3 specialists were assigned to test MD patient group with chinese wechsler intelligence scale for children (C-WISC) and children adaptive capacity scale (CACS). All testing staff was trained collectively for unified standard. C-WISC, developed by Gong, is applicable for children intelligence assessment aged from 6 to 16, including language and operation subscales. Reliability and validity of this scale have been verified strictly, meeting psychometric requirements. Chinese wechsler intelligence scale for adult (C-WISA) was used to assess adults' intelligence. On the other hand, CACS, developed by Zuo, is applicable for children adaptive behavior assessment aged from 6 months to 15 years, covering 6 aspects including independent living, kinetic capacity, homework handling capacity, social interaction, collective activity and self-management. Reliability and validity of this scale also have been verified strictly, meeting psychometric requirements. Adult rating scale for mental disability (ARSMD) was used to assess social function impairment for adults.
Wilcoxon rank sum test was used to compare the expression levels of miRNA between all patient groups and healthy control groups. ROC curve was established to test the sensitivity and specificity of certain miRNA as diagnosis biomarker for psychiatric diseases. All data were processed by SPSS v17.0 (Chicago, IL, USA). P < 0.05 was considered statistically significant.
Wilcoxon rank sum test was used to compare the 10 miRNA expression levels between MDD patient group and control group. The results demonstrated that compared with control group, the expression levels of 5 miRNAs, including miR-26b, miR-4743, miR-4498, miR-4485 and miR-1972 were significantly down-regulated (Table 1).
Table 1: Comparison of miRNA expression between MDD Group and its control group (ΔCT median). View Table 1
Wilcoxon rank sum test was used to compare the 5 MDD related miRNA expression levels between SZ patient group and its control group. The results demonstrated that compared with control group, the expression levels of 5 miRNAs, including miR-26b, miR-4743, miR-4498, miR-4485 and miR-1972 were significantly down-regulated (Table 2).
Table 2: Comparison of MDD related miRNA expression between SZ Group and its control group (ΔCT median). View Table 2
Wilcoxon rank sum test was used to compare the 3 miRNAs (miR-26b, miR-4743 and miR-4485) expression levels between MD patient group and its control group. The results demonstrated that compared with control group, the expression levels of miR-4743 and miR-4485 were significantly down-regulated (Table 3).
Table 3: Comparison of miRNA expression between MD Group and its control group (ΔCT median). View Table 3
Wilcoxon rank sum test was used to compare the 3 miRNAs (miR-26b, miR-4743 and miR-4485) expression levels between GAD patient group and its control group. The results demonstrated that compared with control group, the expression levels of all three miRNAs were significantly down-regulated (Table 4).
Table 4: Comparison of miRNA expression between GAD Group and its control group (ΔCT median). View Table 4
ROC curve was established by using the down-regulation of miR-26b in SZ patients as testing variable and the normal expression of miR-26b in control group as state variables. The results revealed that down-regulation of miR-26b had significant diagnostic value of SZ (Figure 1, AUC = 0.868, P = 0.000) with critical value, sensitivity and specificity being 0.671, 0.721 and 0.950 respectively. These results indicate that using miR-26b with expression above 0.671 as diagnostic standard for SZ has a sensitivity of 72.1% and a specificity of 95.0% (Figure 1).
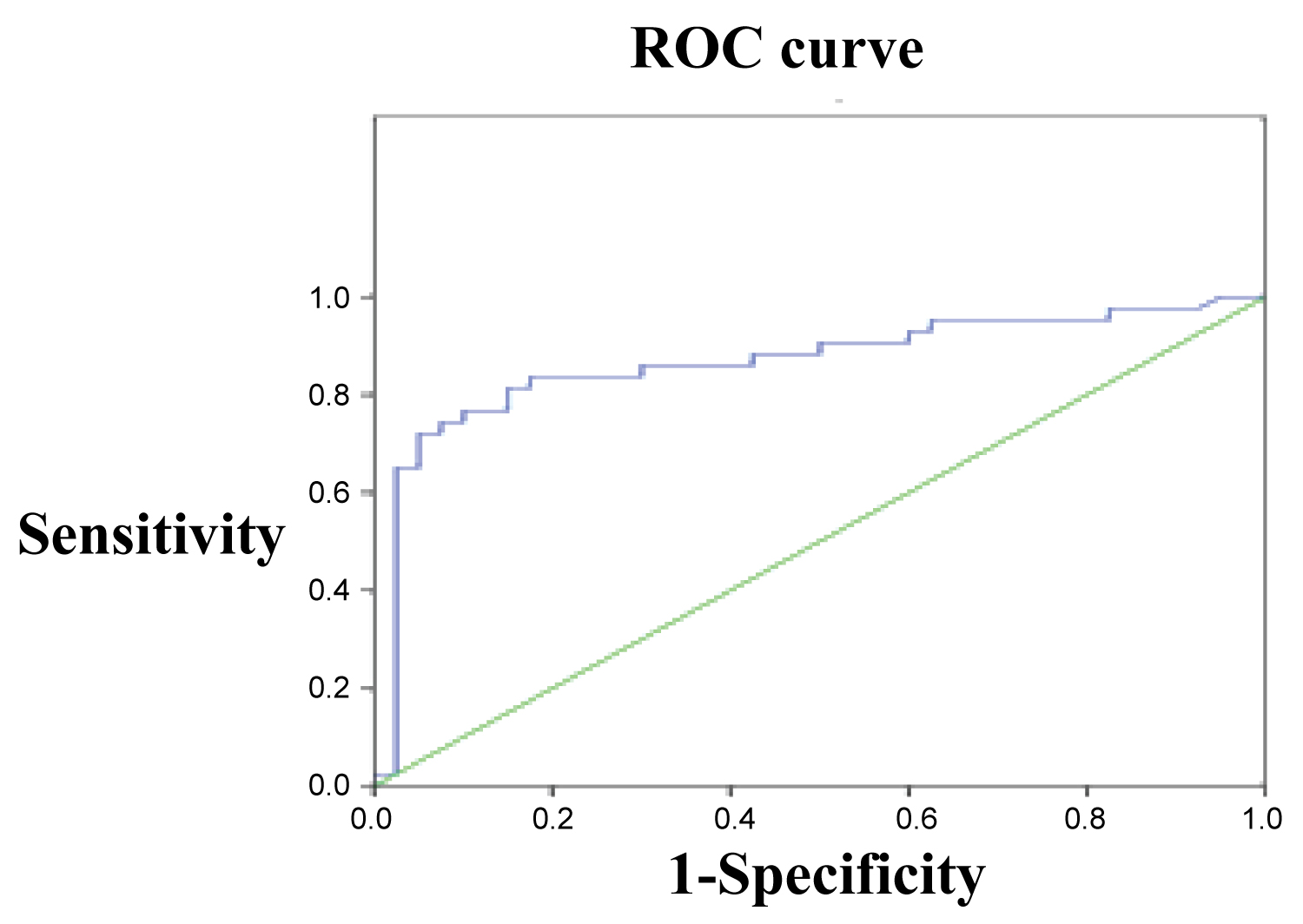 Figure 1: ROC curve of miRNA-26b in SZ group against control group. View Figure 1
Figure 1: ROC curve of miRNA-26b in SZ group against control group. View Figure 1
ROC curve was also established by using the expression of miR-26b in MD patients as testing variable and the expression of miR-26b in SZ patients as state variables. The results revealed that the differential expression of miR-26b had significant value in differentiating between SZ and MD (Figure 2, AUC = 0.765, P = 0.000) with critical value, sensitivity and specificity being 0.455, 0.780 and 0.650 respectively.
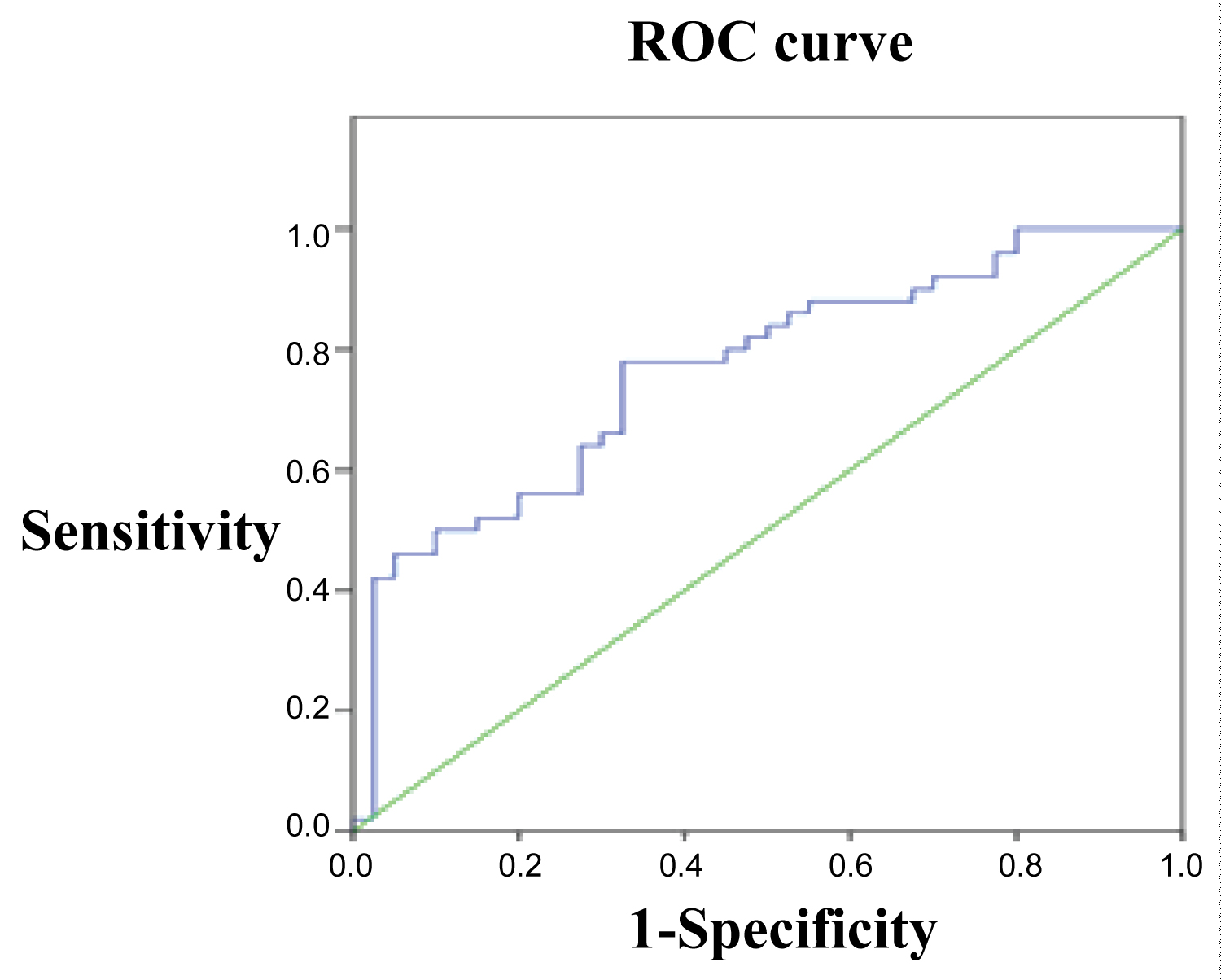 Figure 2: ROC curve of miRNA-26b in SZ group against MD group. View Figure 2
Figure 2: ROC curve of miRNA-26b in SZ group against MD group. View Figure 2
ROC curve was also established by using the expression of miR-26b in GAD patients as testing variable and the expression of miR-26b in SZ patients as state variables. The results revealed that the differential expression of miR-26b had significant value in differentiating between SZ and GAD (Figure 3, AUC = 0.802, P = 0.000) with critical value, sensitivity and specificity being 0.455, 0.667 and 0.825 respectively.
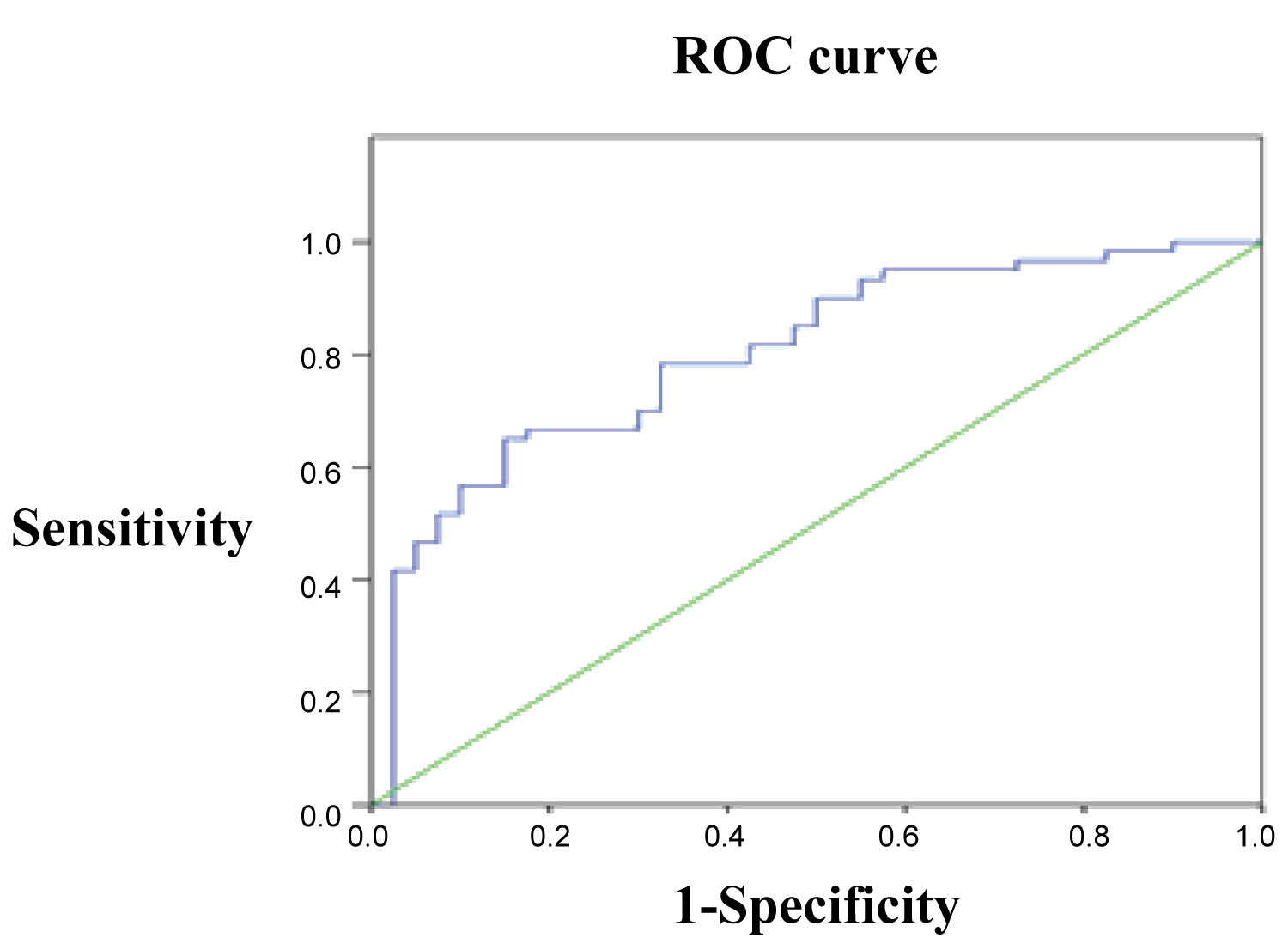 Figure 3: ROC curve of miRNA-26b in SZ group against GAD group. View Figure 3
Figure 3: ROC curve of miRNA-26b in SZ group against GAD group. View Figure 3
ROC curve was also established by using the expression of miR-26b in MDD patients as testing variable and the expression of miR-26b in SZ patients as state variables. The results revealed that the differential expression of miR-26b had significant value in differentiating between SZ and MDD (Figure 4, AUC = 0.629, P = 0.000) with critical value, sensitivity and specificity being 0.212, 0.537 and 0.675 respectively.
 Figure 4: ROC curve of miRNA-26b in SZ group against MDD group. View Figure 4
Figure 4: ROC curve of miRNA-26b in SZ group against MDD group. View Figure 4
Studies on miRNA expression and psychiatric diseases, including SZ, MDD, GAD and others, have been on the increase in recent years. Duan, et al. sampled 2,610 SZ cases and 2,611 controls of European ancestry, and found that rare noncoding risk variants are associated with SZ and BP at MIR137/MIR2682 locus, with risk alleles decreasing MIR137/MIR2682 expression [2]. Song enrolled a total of 20 schizophrenia patients absent of antipsychotics and 20 age-and gender-matched normal controls, and tested for 9 schizophrenia-associated microRNA (miR-30e, miR-34a, miR-181b, miR-195, miR-346, miR-432, miR-7, miR-132 and miR-212) expression levels in plasma using quantitative RT-PCR and for symptomatology improvement using positive and negative syndrome scale (PANSS) before and after treatment, and found that the significant down-regulation of miRNA-181b expression predicts improvement of negative symptoms to treatment, and thus can serve as a potential plasma molecular marker for antipsychotic responses [10]. Fan proved that aberrant miRNA expression was involved into the molecular mechanism of MDD, and might return to normal level after antidepressant drugs treatment [11]. Rett syndrome is a complex neurological disorder that has been associated with mutations in the gene coding for MECP2. A couple of studies have demonstrated that the transfection of miR-146a in a neuroblastoma cell line cause the downregulation of IL-1 receptor-associated kinase 1 (Irak1) levels, suggesting that the identified defect of miR-146a in Rett syndrome mice brains might be responsible for the observed upregulation of Irak1 in this model of the human disease [14-16]. Yet another study used Bioinformatic analysis to identify putative gene targets for these stress-responsive microRNAs, some of which were known to be associated with stress. One of the prominent stress-induced microRNAs found in this screen, miR-34c, was further confirmed to be upregulated after acute and chronic stressful challenge and downregulated in Dicer ablated cells [17,18]. Based upon these studies, we can assume that miRNAs probably serve a significant role in gene modulation and regulation in psychiatric diseases.
In this study, we found that the expression of the 5 identified miRNAs (miR-26b, miR-4743, miR-4498, miR-4485 and miR-1972) in MDD patients [11] were also up-regulated in SZ patients, suggesting the shared clinical symptoms of SZ and MDD, including apathy, lack of euphoria and attention dysfunction and others, may have common molecular genetic basis in terms of peripheral miRNA expression. Among the 3 miRNAs (miR-26b, miR-4485 and miR-4743) differentially expressed in GAD patients, miR-4485 and miR-4743 were also differentially expressed in MD patients. Joel, et al. proved that MD children may usually present symptoms of depression and anxiety [19-21]. Additionally, Wang, et al. found that genotyped 16 SNPs within the CMYA5 gene and performed case-control studies in 1330 schizophrenia patients, 1045 patients with major depressive disorder, and 1235 normal controls, one risk haplotype of rs16877109-rs3828611 (G-G) was associated with both schizophrenia and major depressive disorder. We can infer based upon these studies that every psychiatric disease may find its molecular genetic way back in certain miRNAs expression profiling, and some of psychiatric diseases may share common miRNAs functional pathways.
In order to differentiate diagnostic value of different miRNAs for certain psychiatric diseases, we selected miR-26b abnormally expressed in most of psychiatric diseases as a common marker for ROC curve analysis. The results demonstrated that using miR-26b as biomarker could effectively differentiate between SZ and other three psychiatric diseases, namely MDD, MD and GAD, indicating that miR-26b has significant diagnostic value for SZ.
Caputo's study [22] demonstrated that miR-26a and miR-26b downregulate BDNF expression and that the presence of the variant alleles of two single nucleotide polymorphisms (rs11030100 and rs11030099) mapping in BDNF 39UTR specifically abrogates miRNAs targeting. Furthermore, a high linkage disequilibrium rate between rs11030100, rs11030099 and the non-synonymous coding variant rs6265 (Val66Met), was found, suggesting a hypothesis that miR-26s mediated regulation could extend to rs6265, which proved to be closely involved into many psychiatric diseases. Up-regulation of miR-26b has also been observed in neurons causes pleiotropic phenotypes that are observed in AD [23].
In conclusion, miR-26b has been found to differentially express in many psychiatric diseases, in SZ particularly, and differentiate SZ from other psychiatric diseases. Its neuronal pathology warrants further investigation.
We sincerely thank the patients, their families, and the volunteers for their altruistic participation and services.
Ling-ming Kong & Li-yi Zhang developed the clinical design, collected and organized the participants data, and wrote the first draft of the manuscript, data processing, manuscript polish and revision. Xiao-li Zhu developed the general study design.
None.