Introduction: Hepatitis B virus (HBV) Genotype D has been associated with chronicity leading to Liver cirrhosis and Hepatocellular carcinoma. The Genotype is one of the most common genotypes detected in Nepal. However, the expression analysis for cytokines like IFNγ and IL-6 which could help in boosting the immunity in patients has not been done. The study aims to investigate the expression of IFNγ and IL-6 in chronic HBV patients with Genotype D.
Methods: Sera samples from 43 Hepatitis B infected patients were collected from a tertiary care hospital in Kathmandu, Nepal along with its corresponding control. The information on the patients' age, gender, clinical and biochemical history and controls was collected. Further, serology, copy number detection and genotyping of the selected samples were performed. Lastly, the expression pattern of the IL-6 and IFN-γ was performed on chronic hepatitis B patients with genotype D.
Results: Out of 43 patients who participated in the study, 19 acute patients and 24 chronic patients were detected respectively. The mean age of the patients was 26 ± 6. Further, 33 samples had a viral load of more than 10,000 IU and 10 had a viral load of less than 10,000 IU/ml. Out of 20 Genotype D samples, 18 were positive for chronic HBV. Real-time results of the 18 genotype D samples expressed a 3-fold increase in IL-6 gene relative to GAPDH and IFN-γ showed an 8-fold increase in its expression as compared to the healthy control. Significant statistical difference of p < 0.001 was only considered.
Conclusion: These preliminary results indicate the importance of further studying the relationship between genotypes and cytokines in chronic HBV Infection which could lead to various liver diseases up to HCC. However, the potential role of genotype, fold expression of cytokine IFN-γ and IL-6 in different stages of HBV infection has to be further investigated to better understand the complex interplay of host-pathogen interaction which governs the prognosis of the disease.
Hepatitis B, Genotype, Nepal, Cytokine expression, Chronic hepatitis
The Hepatitis B virus (HBV), a non-cytopathic virus belonging to the Hepadna family is one of the major agents responsible for viral-induced Hepatitis [1,2]. Despite the availability of an effective vaccine, hepatitis B virus (HBV) remains a major public health threat with more than 240 million individuals chronically infected worldwide who are at high risk of developing liver cirrhosis and hepatocellular carcinoma (HCC) [3,4]. The term chronic Hepatitis B infection is defined as persistence of hepatitis b surface antigen (HBsAg) for six months or more [5]. In chronic patients, the Immune reactive phase (HBeAg-positive) is more frequent in individuals infected in adulthood. However, this stage might be followed by the HBV inactive carrier stage (defined by the absence of HBeAg and presence of anti Hbe with serum HBV DNA levels less than 2000 IU/ml) [6]. In both of the above-stated conditions, there are reports of patients unable to clear the virus and hence leading to persistent infection which might further lead to various liver diseases [5,6]. The conversion of the Immune reactive phase to various phases and hence the outcome of the HBV infection is however dependent on the complex interplay of the host-pathogen interaction.
Regarding the virus, Abundant research indicates the relationship of the HBV genotypes with the severity of infection, disease outcome and with response to the therapy [7,8]. Based on the nucleotide divergence of more than 8%, the HBV has been classified into eight genotypes ranging from A to H [9]. Clinically, Genotype D along with A has been associated with more chronicity ratios than genotype, B and C [10]. Also, Genotype A shows more recombination than other genotypes [11]. Similarly, genotype D has been associated with lower sero clearance of HBsAg and higher prevalence of BCP A1762T/G1764A mutation leading to chronic liver disease [12,13].
Similarly, regarding the host, cytokines have been identified as an essential and critical player in Th1 and Th2 response against the pathogen [14]. Among the Th1 cytokine, interferon gamma (IFN-γ) has been associated with HBV infection and an increased level of the cytokine has been linked with the stimulated immune response leading to cure of the disease [15,16]. The proinflammatory cytokine activates the lymphocytes and the CD8+T cells. Further, any changes in its expression can indicate the risk of hepatitis progression [17,18].
Similarly, interleukin 6 (IL-6) has been associated with chronic inflammatory disease including Chronic hepatitis [19]. Reports suggest that IL-6 promotes T helper cell differentiation as well as the differentiation of CD8+ cells into cytotoxic T cells [20,21]. Also, it has been associated with activation of the JAK/STAT3 pathway which in turn activates several cellular signaling processes including STAT1 and STAT3 which comprises one of the key components of the host-pathogen interaction [22,23].
Nevertheless, the Genotype of the Hepatitis B virus, IFN-γ, and IL-6 may play a key role in the host-pathogen interaction which may determine the outcome of infection. Hence based on this information, an attempt was made to study the expression of IFN-γ and IL-6 in Chronic Hepatitis B patients with Genotype D infection from a tertiary care hospital in Kathmandu, Nepal.
We included in this study adult individuals, older than 18 years, with clinical and serological evidence of HBV infection. Two subgroups, such as acute infection and chronic immune tolerant infection, were recruited for the study. For the study, clinical history, serological tests results (HBsAg, anti-HBsAg, HBeAg, anti-HBeAg, anti-HBcAg) were collected for at least 24 months. Moreover, we obtained data about ALT, aspartate aminotransferase (AST), alkaline phosphatase (AlP) to better categorize patients. Lastly, the detection of the copy number of the virus as well as the genotyping of the selected samples was performed. Further, the expression pattern of the IL-6 and IFN-γ was performed. Informed consent and the ethical approval of the study were taken from the nepal health research council (NHRC), Nepal.
The controls were matched with the patients by number, age, gender, HIV, and HCV status.
Patients were included in this study if their age was over 18 years, and their clinical history included at least two time points of observation separated by at least 6 months for at least 24 months.
Controls were included in this study if they were older than 18 years with no evidence of HBV infection (defined as negative HBsAg, HBeAg, and anti-HBcAg IgM and IgG, with no history of HBV vaccination).
Patients were excluded when they were younger than 18 years, more than one case in the same family, use of immunosuppressive drugs, coinfection with human immunodeficiency virus (HIV) or hepatitis C virus (HCV).
Controls were excluded if they were younger than 18 years, related to patients, or they used immunosuppressive drugs.
Serological screening of the blood samples for HBsAg, HBeAg, Anti-HBsAb, Anti HBe, and Anti-HBc was performed using a commercial kit Tulip Diagnostics (P) Ltd (Altosantacruz, Bambolim complex, Goa, India) as according to the manufacturer's protocol. Further, the strong positive and/or negative were considered for further investigation.
The viral DNA extraction was performed by QIAamp DNA mini kit (Qiagen, Hilden Germany) according to the manufacturer's instructions. Briefly, 200 μl of the serum was mixed with 20 μl of Proteinase K along with 200 μl of the buffer. The whole mixture was vortexed for 15 sec and incubated at 56 °C for 10 min. After brief centrifugation, 200 μl of ethanol was added and the eluent was collected in the collection tube. This step was repeated using the buffer AW1 and AW2. Lastly, AE buffer was added, incubated at room temperature for 1 minute, and then centrifuged at 8000 rpm to collect the final eluent containing the viral DNA. 30 μl of the eluent was collected. 20 μl was used for the genotyping and the remaining 10 μl was stored at -80 °C. The purity and concentration of isolated DNA were evaluated on nanodrop. The good quality of DNA obtained (A260/A280 = 1.7 to 2.0) was further selected for viral load estimation.
The viral load was detected by using Arthus Step one Real-time PCR Kit (Qiagen, Hilden, Germany). Briefly, the internal control was added during sample preparation. Further, 25 μl of the master mix was added to each required well. To this, 25 μl of the sample followed by 25 μl of the quantification standard, Positive control, and Negative control were added in respective wells. The PCR temperature profile for the viral load was Denaturation of 95 °C for 2 mins followed by 0.15 mins and then the amplification at 58 °C for 0.20 mins and then 72 °C for 0.30 mins.
Further for genotype determination, Real-time PCR genotyping was performed using Sacacae Real TM PCR kit (Como, Itali) according to manufacturer's instructions. Briefly, 15 μl of the PCR mix was added with 0.5 μl of hot-start Taq Pol. To this mixture, 10 μl of the DNA was added. Further, 10 μl of Positive Control, as well as negative controls, were added in the well and the reaction conditions were 95 °C for 15 minutes, 95 °C for 5 sec, 60 °C for 20 sec, 72 °C for 15 sec, 95 °C for 5 sec. Further, the fluorescence was obtained at 60 °C for 20 sec and 72 °C for 15 sec.
The RNA extraction was performed according to the manufacturer's instruction using Direct-ZolTM RNA Miniprep (Zymo Research, Tustin, U.S.A). Briefly, 100 μl of the sera sample lysed in TRI Reagent was added with the equal volume of ethanol. The mixture was transferred into a spin column in a collection tube and then centrifuged. The column was transferred into a new collection tube and the flow-through was discarded. 400 μl of the RNA wash buffer was then added into the column and centrifuged. Further, 400 μl of RNA prewash was added to the column and centrifuged. The step was again repeated. Now, 700 μl of RNA Wash buffer was added to the column and centrifuged for 1 minute to ensure removal of wash buffer. Finally, RNA was eluted by adding 50 ul of DNase/RNase-Free water to the column matrix and centrifuging it. The eluted RNA was stored at -20 °C for further use.
The c Dna preparation was done according to the manufacturers instruction using QuantiTectR Reverse Transcription kit (Qiagen, Hilden, Germany)-Briefly, genomic DNA elimination reaction comprising of 2 μl g DNA wipeout buffer, Template RNA and RNAse free water was incubated for 2 minutes at 42 °C. Further, the above mixture was added with 1 μl reverse transcriptase, 4 μl RT buffer and 1 μl RT primer mix and incubated for 15 minutes at 42 °C. Lastly, the mixture was incubated for 3 minutes at 95 °C to inactivate Reverse transcriptase.
The expression analysis of IL6 and IFN-γ was performed by SYBR Green real time PCR method. The results were normalized with the expression level of GAPDH as the house keeping gene. The primer sequences for amplification of IL 6, IFN-γ and GAPDH were GAPDH F: 5'GTCGCTGTTGAAGTCAGAGG-3' GAPDH R: 5'-GAAACTGTGGCGTGATGG-3'; IL 6 F: 5'-AATTCGGTACATCCTCGACGG-3' IL 6 R:5'-GGTTGTTTTCTGCCAGTGCC-3' IFNG F: 5'-CGGAATTCATGTGTTACTGCCAGGAC-3' and IFNG R:5'- CGGGATCCTTACTGGGATGCTCTTCG-3'. The delta ct in the control group were calculated and then the delta ct in HBV patients were calculated. Further, the fold increase in expression was calculated using livak (2delta delta ct) method [24].
The statistical software (IBM/SPSS statistic version 19) was used for data analysis and p-value of less than 0.05 was considered significant.
We enrolled 43 patients (19 acute and 24 chronic patients) and an equally matched control with a mean age of 26 ± 6 and 27 ± 8 respectively, 13(30.33%) of them were females and 30(69.76%) were males. The serology of the patients was HBsAg+/HBeAg+/AntiHBsAb-/AntiHBeAb-and or AntiHBc±. However, regarding viral load, 33(76.74%) samples of viral load more than 10,000 IU and 10(23.22%) samples having viral load of less than 10,000 IU/ml were detected (Table 1).
Table 1: Demographic, serology, and copy number details of the subjects. View Table 1
The patients with viral load more than 10,000 IU/ml were further tested for genotyping. Out of 33 samples tested for genotyping, the genotypes detected are given in Table 2.
Table 2: Shows the number of the detected genotypes. View Table 2
Further, it was observed that out of 20 Genotype D samples, 18 of the samples were positive for chronic group of patients.
The real time Results of the 18 Samples which were detected as genotype D showed that IL-6 gene relative to GAPDH expressed 3-fold increase in the chronic patients as compared to the Healthy control (Figure 1). Further, IFN-γ showed an 8-fold increase in its expression compared to the healthy control. Significant statistical difference of p < 0.001 was only considered (Figure 2).
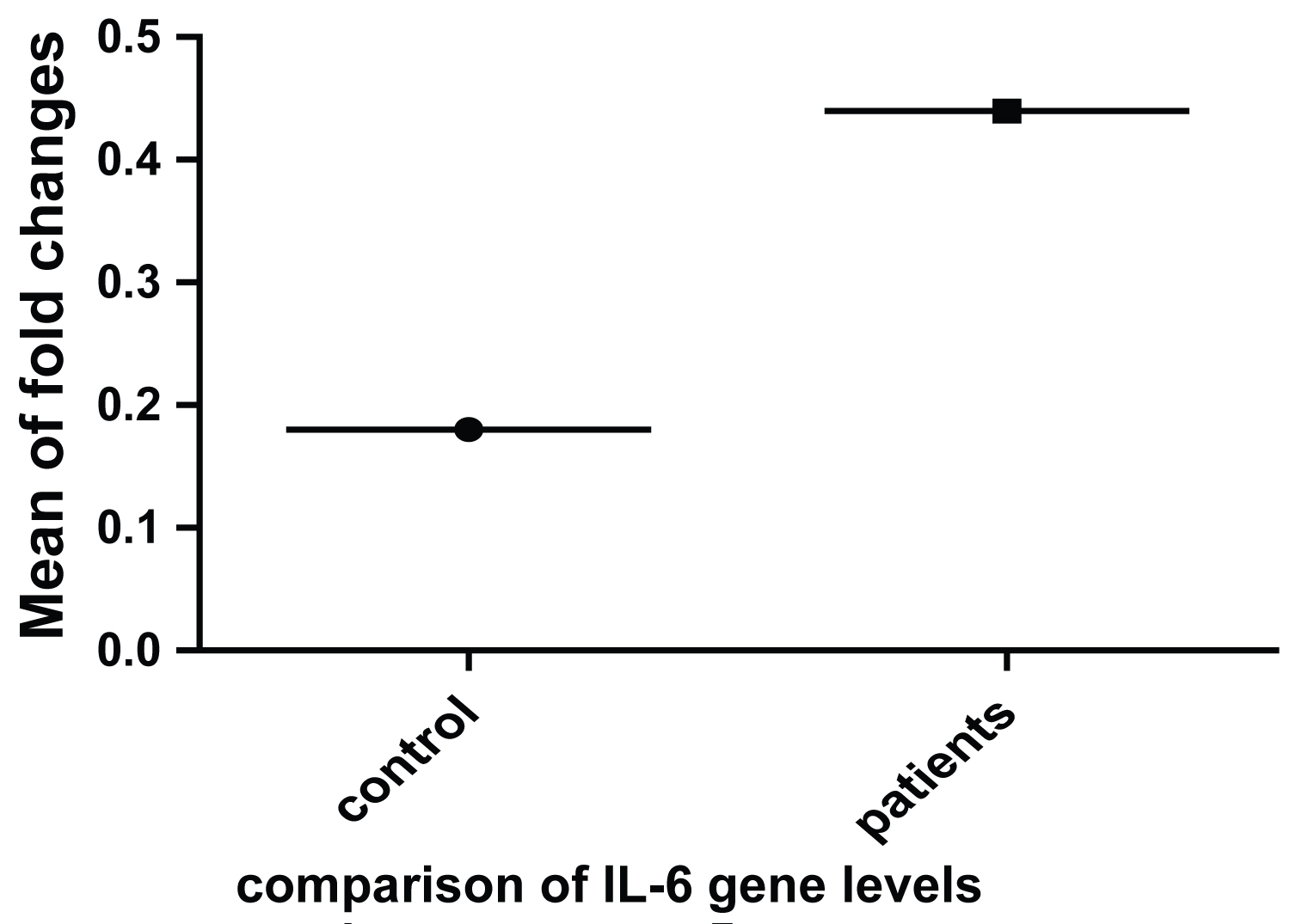 Figure 1: Relative fold changes of IL-6 in HBV patients as compared to healthy controls.
View Figure 1
Figure 1: Relative fold changes of IL-6 in HBV patients as compared to healthy controls.
View Figure 1
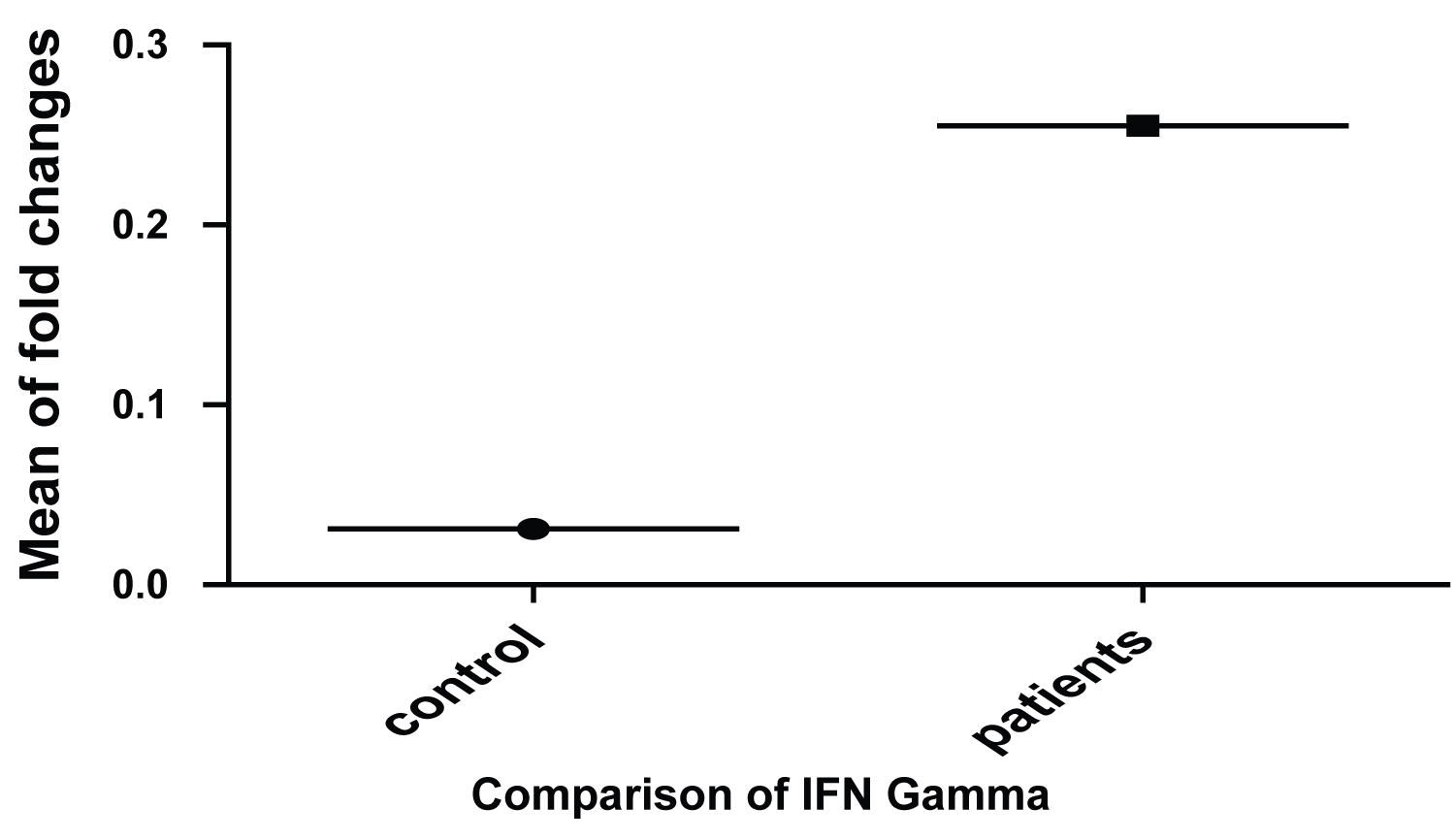 Figure 2: Relative fold changes in the expression of Interferon-γ in HBV patients as compared to its healthy controls.
View Figure 2
Figure 2: Relative fold changes in the expression of Interferon-γ in HBV patients as compared to its healthy controls.
View Figure 2
Hepatitis B virus is a diverse disease with varied prognosis and clinical outcomes. According to the WHO fact sheet, 257 million people have been reported to be infected with chronic hepatitis B infection with an estimate of 887 000 deaths from cirrhosis and hepatocellular carcinoma [25]. The effective management and treatment of the hepatitis B infection is dependent on varied molecular and immunological facets determined in discrete stages of the infection. In such a scenario boosting the host Immune system could help in the control and management of the disease.
Genotyping has been frequently associated with different clinical outcomes and management of the infection [26]. Similarly, the immunological aspect involves the cytokines as an important immunomodulatory molecule that play a salient role in the management and treatment of the disease. Studies have shown that Th1 and Th2 cytokines may contribute to successful treatment of the hepatitis B infection and may be associated with persistence of the infection respectively [27,28]. IL 6 and IFN-γ have been associated with tissue damage, infection, and chronic progression of hepatitis [1,29]. IL 6 is a Th2 cytokine that has been associated with both pro and anti-inflammatory effects and hence plays a crucial role in the activation and regulation of immune response [30,31].
Based on the above studies, an investigation on the expression of IFN-γ and IL-6 was performed on the Chronic Hepatitis B positive patients with Genotype D. Genotype D has been known to remain in inactive carrier state in most cases but a small portion of it might lead to Liver cirrhosis and then Hepatocellular carcinoma [32,33]. Our results stated that the majority of the genotype detected in the study was Genotype D which might effect the prognosis and management of the infection. The work supports the previous research of the genotyping in Nepal where Genotype D is detected as one of the common genotype distributed in Nepal along with Genotype A [34,35].
Various studies have indicated IFN-γ as a cytokine which is responsible for inhibition of HBV replication through non cytolytic process and in showing its impact in the etiology of acute inflammation [2]. Our results suggested that IFN-γ expressed itself in 8 folds as compared to healthy controls. Similar work is supported by Wang, et al. and Seyed, et al. where the concentration of IFN-γ was higher in patients as compared to the healthy populations [36,37]. No significant correlation was found between the cytokines. Similarly, 3 fold increase in IL- 6 expression as compared to healthy controls indicated its association with the Hepatitis B virus infection. IL-6 have been associated with Autoimmune Hepatitis and increase levels of IL-6 have been observed in chronic Hepatitis B positive patients whose condition later progressed to cirrhosis hence indicating its role in chronic hepatitis [38]. Besides this, T tests further validated that the delta ct for both the genes between the hepatitis B positive patients and healthy subject is statistically significant.
In conclusion, Hepatitis B positive chronic patients with genotype D expressed 8-fold increase in IFN Gamma and 3-fold increase in IL-6. Thus, the fact can be used as an aid for preventing the viral progression by better understanding the roles of the genotype, cytokine IFN-γ and IL-6 for the use of various antiviral drugs and further for the development of a novel Immunotherapeutic strategy for Hepatitis B virus infection. Hence, our findings may help in improvement in the clinical treatment and management of patients with the infection. However, other aspects of the Immune response of this disease need to be further investigated.
We acknowledge University Grants Commission for providing partial funding for the research.
The Authors disclosed receipt of the following finanacial support for the research. The research was partially supported by university grants commission (UGC), Nepal.
The authors state no conflict of interest for the research.
Informed consent and the ethical approval of the study were taken from the nepal health research council (NHRC), Nepal (Ref Number: 138/2018).