The advent of nanotubes in the nanotechnology world has led to significant advances in a number of biological and materials application, due to their structural properties such as the surface to volume ratio and potential to surface carrying or inside capsulation any materials. Among the various types of nanotubes, DNA nanotubes, due to their unique characteristics, such as precis controllability and programmability in shape/size/length/diameter and its biological origin compared to different types of nanomaterial, can be the suitable candidate for template patterning alignment and precise organization of nanoparticles at surface or into channel. These characteristics can be used in nanoelectronic devise or in the field of diagnostic nanobiosensores.
Here, we report a new construction methodology for encapsulation of magnetic nanoparticles inside DNA nanotubes channel. Constructed-simultaneously encapsulation of magnetic nanoparticles into the large channel of this tubes leads to "pea-pod" particle alignment in nanotube channel.
Transmission electron microscopy and atomic force microscopy confirmed the fabrication of DNA nanotubes contained the magnetic nanoparticles inside the channel.
These biohybrid nanomaterial would be proposed as the nanoarray platform in nanobiosensing devices.
DNA nanotubes, Magnetic nanoparticles, Transmission electron microscopy, Atomic force microscopy, Nanobiosensors
Deoxyribonucleic acid (DNA) has recently suggested opportunities for the fabrications of nanostructures and massively construction of the artificial nanoarchitectures with various geometries or patterns by the self-assembly phenomena [1-6]. Scaffolded DNA origami has become a unique technique for designing well-defined nanostructures with any desired shape and pattern because of programmability and controllability in fabrication and for the controlled arrangement of functional nanostructures with few nanometer resolutions [7,8]. DNA nanotechnology also has used DNA strands to assemble gold, Magnetic and other nanoparticles into distinct 1, 2-D structures, or into periodic 3-D crystals [9,10]. Combining the novel electronic, magnetic, and optical properties of either semiconductor or metallic nanoparticles with the self-assembly properties of DNA strands can led to construction of new hybrid bio-nanomaterial with ability to precise organization and alignment of nanoparticles [11,12]. These unique features make DNA origami nanostructures appropriate candidates for use as platform to alignment of nanoparticles for nanodevices fabrication. Consequently, a number of studies have shown the precise organization of metallic nanoparticles on/in various DNA origami shapes [13,14].
Among the various types of DNA nanostructures, DNA nanotubes, due to their unique characteristics, such as surface-to-volume ratio, controllability and programmability in size and fabrication, ability to carry material over the surface, encapsulation of material inside the channel and its biological origin compared to different types of nanomaterial, has a vast potential for application such as patterning to precise organization of nanoparticles that can be used as a sensing platform in the field of diagnostic nanobiosensors [15-17]. In fact, DNA nanotubes are filamentous structures that are made from double-stranded DNA helix by bottom-up or top-down techniques with a hollow central channel, and walls made of DNA [18].
The most important applications of DNA nanostructures coupled with nanoparticles are their ability to arrange some materials on them, which directly relate to the programmability of DNA molecules. These nanostructures could serve as the excellent platforms for carrying nanoparticles, antibodies, and protein while the functional groups were incorporated inside and outside of the nanomaterials [19-23]. Periodic array of nanoparticles on surface or in channel of DNA nanotubes explored their potentials for the several applications such as sensing platform in nanobiosensors or DNA nanowire fabrication [15-17]. The fabrications of DNA nanotubes particularly with the incorporations of 1-D nanoparticles such as metallic and magnetic nanoparticles have expanded their applications in nanotheraputics and nanodiagnostics [5,6,19,20]. Physical properties of the nanocomplexes change with the incorporation of the nanoparticles within the DNA nanotubes [21,22].
The improvement of any aspect of these physical characteristics could greatly expand the range of employing theses nanoarchitectures in various areas of the science and technology [15-23].
DNA nanotubes could be incorporated with the nanoparticles through the non-covalently binding to the positively charged nanoparticles; however, for achieving the surface patterning or positioning of DNA nanotubes by nanoparticles could also being possible by covalently attaching the functional groups or molecules [24,25].
In this study we described a new procedure for the origami induced alignment of cationic-charged magnetic nanoparticles inside DNA nanotubes (Figure 1). The incorporation of the positively charged nanoparticles in the DNA nanotubes can be occurred simultaneously within a DNA-origami self-assembly reaction. By this methodology, DNA nanotubes can be suggested as nanocarriers; however, some novel physicochemical properties would be investigated from the nanotubes due to their carrying nanoparticles.
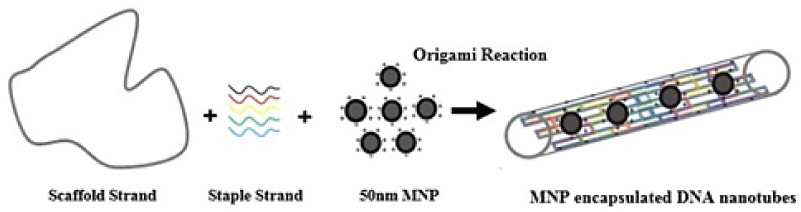 Figure 1: Schematic graphical view of DNA nanotube filled up with the magnetic nanoparticles.
View Figure 1
Figure 1: Schematic graphical view of DNA nanotube filled up with the magnetic nanoparticles.
View Figure 1
Theses nanotubes were characterized by transmission electron and atomic force microscopes. Also, their electrophoretic nobilities were studied using the gel electrophoresis experiment.
The cadnano software was used for designing desired DNA nanotubes in this study. DNA sequence of M13mp18 phage genome was applied in the software as the scaffolded strand to achievement of oligonucleotides sequences of DNA staple as a helper for self-assembly of scaffolded strand into DNA nanotubes. The staple strands sequence was determined based on their complementary with the special site of scaffold that bind to form desired structure. The honeycomb style was introduced in the cadnano for properly folding of DNA nanotubes within a DNA origami reaction.
The origami reaction was prepared by mixing 20 nM DNA scaffold M13mp18 single-stranded DNA (from New England Biolabs; Massachusetts, USA) with 100 nM of each staple oligonucleotide (synthesized and desalted by Bioneer; Korea) that those were prepared in 1X Tris, acetic acid, and EDTA buffer (40 mM Tris-acetic acid buffer, pH 8.0, and 12.5 mM magnesium acetate). The mixture was kept at 95 ℃ for 5 min for denaturation of undesired secondary structures of any strand and between them and then cooled from 95 ℃ to 20 ℃ with a constant rate of -1 ℃/min in the Rotor Gene Q thermocycler (QIAGEN, Germany) for annealing of desired shape. For the fabrication of long and sturdier DNA nanotubes, the origami products were treated by ligation process with T4 DNA ligase. The ligation-reaction mix was prepared containing 2 µL of 10X T4 DNA ligase reaction buffer, 10 µL of self-assembled DNA nanotubes, 2 µL of 50% polyethylene glycol, and 1 µL of T4 DNA ligase enzyme. Finally, the ligation mix was incubated at 37 ℃ for 1 hour.
For fabrication of DNA nanotubes including magnetic nanoparticles inside the channel, cationic-charged MNPs (fluidMAG-UC/C: Cationic charged, with 50 nm hydrodynamic dimension measured by the manufacturer (Chemicell GmbH, Germany) with dynamic light scattering; DLS) were used with the hypothesis that these MNPs have tendency to physical adsorption to DNA strands and was stabilized by cross-linking with the internal negative charge of DNA nanotube resulting from anionic phosphate group. By considering it, fabrication of DNA nanotube by top-down method was accompanied and joined by including process of cationic charged MNP (C-MNP). For this purpose, 2.5 × 10-3 mg/ml C-MNP (from Chemicell GmbH, Germany) was combined with the Origami reaction mix of DNA nanotube. The origami reaction was prepared by mixing 20 nM DNA scaffold M13mp18 single-stranded DNA (from New England Biolabs; Massachusetts, USA) with 100 nM of each staple oligonucleotide (synthesized and desalted by Bioneer; Korea) that those were prepared in 1X Tris, acetic acid, and EDTA buffer (40 mM Tris-acetic acid buffer, pH 8.0, and 12.5 mM magnesium acetate). The mixture was kept at 95 ℃ for 5 min and then annealed from 95 ℃ to 20 ℃ with a constant rate of -1 ℃/min in the Rotor Gene Q thermocycler (QIAGEN, Germany). For the fabrication of long and sturdier DNA nanotubes, the origami products were treated by ligation process with T4 DNA ligase. The ligation-reaction mix was prepared containing 2 µL of 10X T4 DNA ligase reaction buffer, 10 µL of self-assembled DNA nanotubes, 2 µL of 50% polyethylene glycol, and 1 µL of T4 DNA ligase enzyme. Finally, the ligation mix was incubated at 37 ℃ for 1 hour. Then, the fabricated magnetic DNA nanotubes were separated from simple DNA nanotubes and others by Magneto PURE-Micro separator (from Chemicell GmbH, Germany).
For assessing the electrophoretic behaviors of these nanotubes filled-up with magnetic nanoparticles, 5 µL of the nanotubes were loaded in 1% agarose gel and the assay was run within an electrophoresis experiment at 0.5X TBE buffer pH 8.2. The experiment was performed using an electrophoresis mini set from Bio-Rad (USA). The electrophoresis behaviors of the nanotubes were compared with standard natural DNA (i.e., GeneRuler DNA Ladder Mix, Thermo Fisher Scientific, Inc., Waltham, MA, USA). The gel was stained according to the manufacturer instruction (SYBR Gold nucleic-acid gel stain, from Molecular Probes Inc., Eugene, Oregon, USA).
Purification and Extraction of Magnetic DNA nanotubes from electrophoresis gel Extraction of DNA nanotubes from gel is required for purification of DNA origami products from unwanted nanostructures. For extraction of DNA nanotubes from the gel, "Quantum Prep Freeze N Squeeze DNA Gel-Extraction Spin Column, as a gel-specific kit from Bio-Rad (USA) was used". Initially, using a clean razor blade, carefully excise the bond of interest. Trim excess agarose from all six sides of the DNA nanotubes bond to maximize recovery and purity. Then the trimmed gel slice was chopped and placed the pieces in the filter cup of dolphin shaped microtube in -20 ℃ freezer for 5 minutes. The slice was then centrifuged in a gel extraction column with a proprietary tool and centrifuged at 13000 g for 3 minutes. The final product obtained from gel extraction, used as tubular DNA nanotubes incorporated of magnetic nanoparticles, was used for characterization studies.
Transmission electron microscope (TEM) was used to assess the size and morphology of magnetic DNA nanotubes and also the simple DNA nanotubes. For this purpose, the DNA nanotubes were immobilized by syringe spraying on Agar Scientific (Stansted, Essex CM24 8GF, United Kingdom) from carbon layer with 300-mesh Cu (50). The micrographs were obtained by TEM EM900 (Zeiss, Germany). Topographic characteristics of the nanotubes were studied by atomic force microscope (JPK-AFM, Germany). For this purpose, 5 µL of the nanotubes was immobilized on a mica surface for 4 hours at 25 ℃. The AFM was performed in contact mode with JPK-AFM (with 150 Hz IGain, 0.0048 PGain, and 1.0 V set point via a JPK NanoWizard control). The cantilever was ACTA-10 probe model. The rough data were analyzed using the JPK Nano analyzer software.
The computational design of the nanotube from cadnano software was indicated in Figure 2.
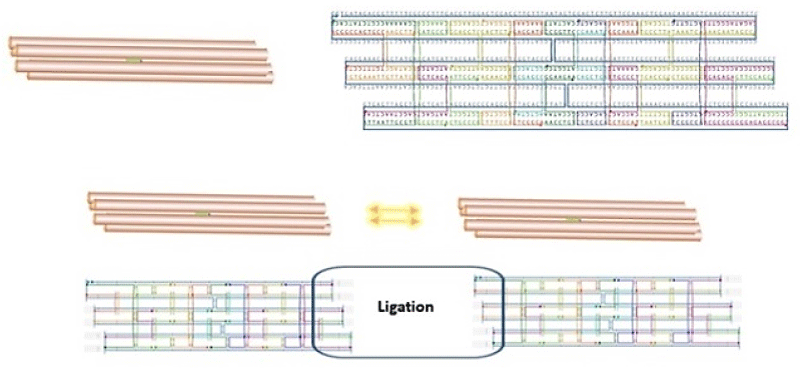 Figure 2: Schematic graphical view of DNA nanotubes designed in this study by cadnano.
View Figure 2
Figure 2: Schematic graphical view of DNA nanotubes designed in this study by cadnano.
View Figure 2
This platform showed a cylindrical scaffold assembled with the staples. The scaffold with 6 single-stranded DNA of M13mp18 nucleotide sequences that those were marked with blue color. The sequences of staples were determined as 24 oligonucleotides (Table 1) that those were marked with different colors in Figure 2.
Table 1: Oligonucleotide staples. View Table 1
TEM micrographs of DNA nanotubes without and included of magnetic nanoparticles were shown in Figure 3 and Figure 4.
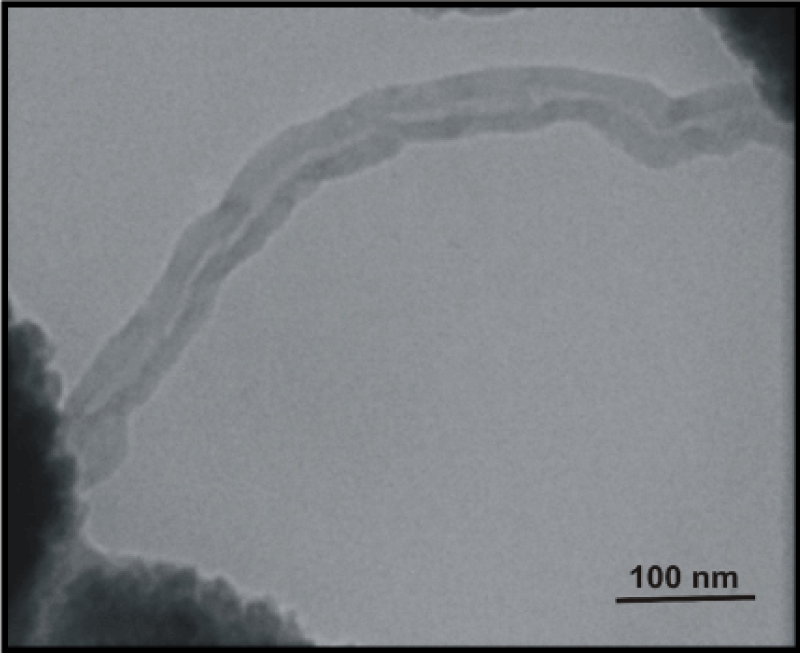 Figure 3: TEM micrograph of DNA nanotubes without of MNP.
View Figure 3
Figure 3: TEM micrograph of DNA nanotubes without of MNP.
View Figure 3
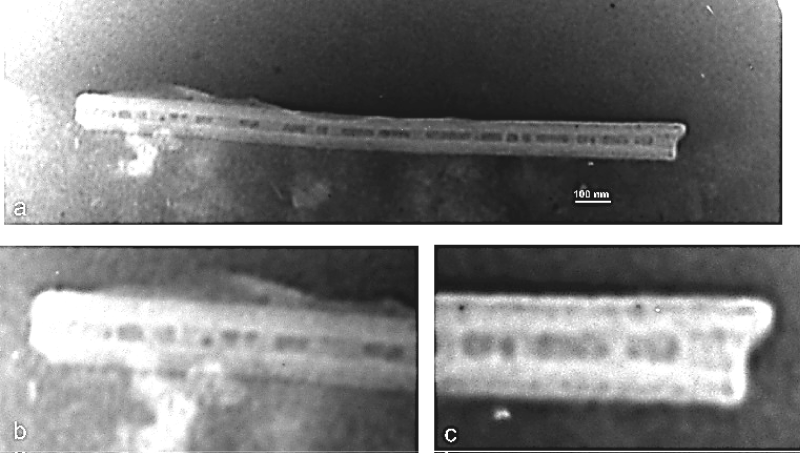 Figure 4: a) TEM micrograph of DNA nanotube contained MNPs; b,c) Maximized ends of the nanotube shown at 'a' image that those demonstrated MNPs densely in the channel of DNA nanotube.
View Figure 4
Figure 4: a) TEM micrograph of DNA nanotube contained MNPs; b,c) Maximized ends of the nanotube shown at 'a' image that those demonstrated MNPs densely in the channel of DNA nanotube.
View Figure 4
AFM results also demonstrated the nanoparticle-filled nanotubes (Figure 5); however, the free C-MNPs among DNA nanotubes were also seen that those were not encapsulated within the nanotubes. Constructed-simultaneously encapsulation of magnetic nanoparticles into the large channel of this tubes leads to "pea-pod" particle alignment in nanotube channel which is visible in AFM topography view.
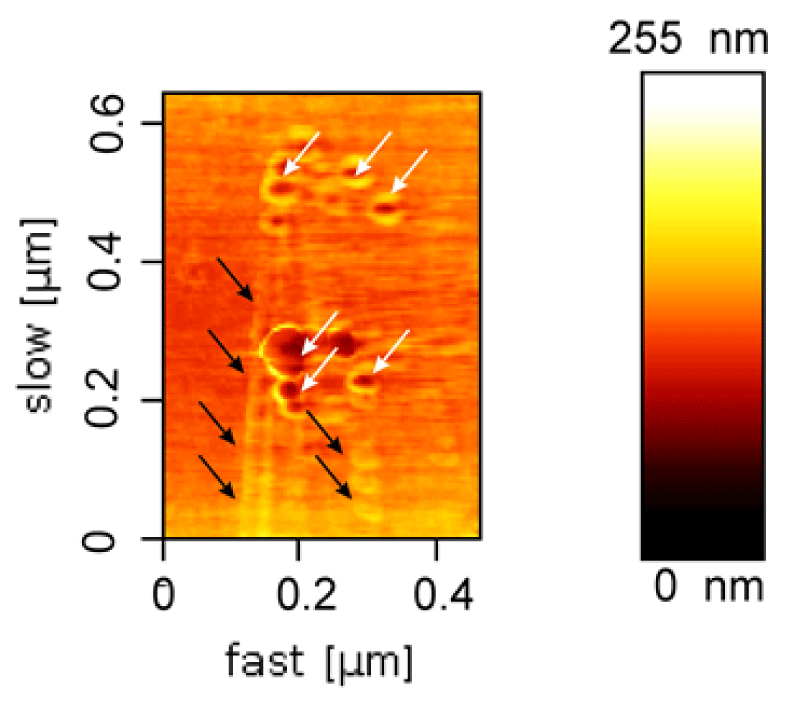 Figure 5: AFM of DNA nanotubes filled up with an array of C-MNPs (black arrows). Also, a few magnetic nanoparticles outside of DNA nanotubes were indicated (white arrows).
View Figure 5
Figure 5: AFM of DNA nanotubes filled up with an array of C-MNPs (black arrows). Also, a few magnetic nanoparticles outside of DNA nanotubes were indicated (white arrows).
View Figure 5
The electrophoretic behavior of DNA nanotubes contained C-MNPs was reversed in comparison to that behavior by natural DNA (e.g. DNA ladder) (Figure 6).
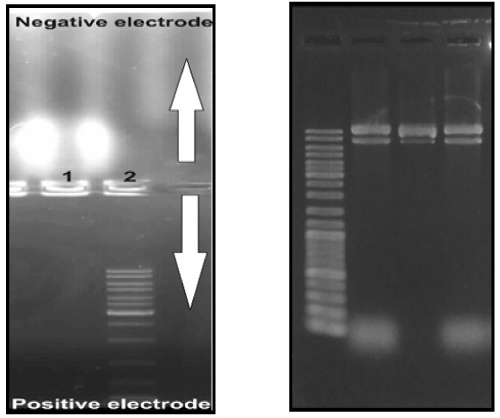 Figure 6: Electrophoretic movement of DNA nanotubes without and contained C-MNPs respectively (right and Left) in comparison to DNA ladder.
View Figure 6
Figure 6: Electrophoretic movement of DNA nanotubes without and contained C-MNPs respectively (right and Left) in comparison to DNA ladder.
View Figure 6
The nanoparticle-filled up nanotubes indicated positively charges and those moved toward the negative pole, whereas the natural DNA with the negatively charges moved to the anode. This behavior from the magnetic DNA nanotubes confirmed successfully the encapsulation of the cationic-charged MNPs by DNA nanotubes within their fabrication process.
The principles of DNA nanotubes design and fabrication used in this research were reported previously [18,26,27]. Here 24 staple sequences were determined for properly folding DNA nanotubes in the presences of cationic charged magnetic nanoparticles. The electrophoretic behavior of DNA nanotubes was previously studied. Simple DNA nanotubes showed the migration similarly to natural DNA from negative pole (cathode electrode) to positive pole (anode electrode) due to their negatively charges from the phosphate groups on their surfaces. While, the DNA nanotubes contained cationic charged magnetic nanoparticles fabricated in this study showed different electrophoresis behavior when compared with that by the natural DNA (Figure 5). Magnetic DNA nanotubes indicated positively charges due to the encapsulations of the cationic charged nanoparticles and their movements toward to the negatively pole, whereas the natural DNA and the empty DNA nanotubes have been showed migrations from the cathode electrode to the anode electrode within the electrophoresis experiment. The electrophoretic mobility of the magnetic DNA nanotubes suggested the encapsulations of the cationic-charged MNPs by DNA nanotubes; however, the AFM micrograph (Figure 4) demonstrated that the nanoparticles have being encapsulated inside the nanotubes, because in AFM image, it's obvious the periodic array or pea-pod alignment of magnetic nanoparticles in DNA nanotubes channel. In addition, it seemed that the addition of the nanoparticles in the origami reaction could potentially affect the fabrication of the magnetic nanotubes.
In conclusion, we have constructed nanoparticle-filled up DNA nanotubes via a DNA origami reaction contained cationic magnetic nanoparticles. DNA self-assembly was employed via the Origami reaction for obtaining well oriented DNA nanostructures with the affordable precision. The top-down approached fabrication would able to organize self-assembling the nanotubes with the hierarchically defined platforms. By incorporating the other DNA based molecules such as DNAzymes, aptamers, or DNA walkers, the nanotubes could be employed for a wide range of the applications in biomedicine [28-34]. For instance, these DNA nanotubes would be employed for the development of the high-throughput DNA biosensors. This work presented herein using a procedure for filling-up DNA nanotubes with the cationic charged magnetic nanoparticles suggested as the hybrid nanomaterial for carrying biomolecules [35].
There are no conflicts to declare.
University of Isfahan (grant No. A/40284/94) and Mazandaran University of Medical Sciences (grant no. 1271/93) were supported this study. The authors thank also Central Research Laboratory, Mazandaran university of Medical Sciences and Iran Nanotechnology Initiative (INI).
A.R. and P.G. conceived and designed the experiments; A.R. performed the experiments; P.G. and A.R analyzed the data; A.Z. and P.G. contributed reagents/materials/analysis tools; A.R. and P.G. wrote the paper.