International Journal of Oral and Dental Health
Prevalence and Genotypes of Candida albicans from Necrotising Periodontal Disease and the Tongue
Elaine Blignaut1* and Reality Phiri2
1Department of Oral Pathology and Oral Biology, School of Dentistry, University of Pretoria, Pretoria, South Africa
2Department of Prosthodontics, School of Oral Health Sciences, Sefako Makgatho University, Medunsa, South Africa
*Corresponding author:
Elaine Blignaut, Department of Oral Pathology and Oral Biology, School of Dentistry, University of Pretoria, Pretoria, South Africa, Tel: +27-12-319-2320, Fax: +27-12-321-2225, E-mail: Elaine.blignaut@up.ac.za
Int J Oral Dent Health, IJODH-3-038, (Volume 3, Issue 1), Research Article; ISSN: 2469-5734
Received: August 05, 2016 | Accepted: February 09, 2017 | Published: February 13, 2017
Citation: Blignaut E, Phiri R (2017) Prevalence and Genotypes of Candida albicans from Necrotising Periodontal Disease and the Tongue. Int J Oral Dent Health 3:038. 10.23937/2469-5734/1510038
Copyright: © 2017 Blignaut E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Objectives: The objectives were to determine the prevalence of Candida albicans in lesions of necrotising periodontal disease (NPD) and on the tongue of patients and to compare the fingerprinting patterns of C. albicans isolates from the two oral sites.
Methods: Microbiological specimens were taken from NPD lesions and the tongue of 87 patients and cultured on Sabouraud Dextrose and CHROMagar, followed by treating patients according to a standard protocol. DNA fingerprinting, using restriction fragment length polymorphism (RFLP) with a 32P-labelled Ca3 probe, was performed on paired isolates of C. albicans simultaneously isolated from NPD lesions and the tongue.
Results: C. albicans was isolated from 47 (54%) patients in total, from only the tongue in 39 (44.8%), simultaneously from the tongue and diseased sites in 7 (8.0%) and only from diseased site in 1 patient (1.1%). The DNA fingerprinting patterns of the isolates were similar within each pair but differed between the 7 pairs, without any evidence of a predominant genetic subtype among the isolates. The clade affiliation resembled that of previously fingerprinted isolates obtained from the region. No statistically significant correlation was demonstrated between the extent (p = 0.4621) or severity (p = 0.3365) of NPD lesions and the presence of yeasts (Fisher's Exact Test). NPD in all patients presenting for a follow-up visit had resolved with conventional treatment and without the addition of antifungal agents.
Conclusion: No association between C. albicans and NPD could be demonstrated and evidence is presented that it is unlikely that the C. albicans isolated from NPD represent a pathogenic sub-group and are more likely to be contaminants from elsewhere in the mouth.
Keywords
Candida albicans, Ca3 fingerprinting, Necrotising ulcerative gingivitis, Necrotising ulcerative Periodontitis, Necrotising periodontal disease, South Africa, HIV
Abbreviations
NPD: Nectrotising Periodontal Disease; NUG: Nectrotising Ulcerative Gingivitis; HIV: Human Immunodeficiency Virus; RFLP: Restriction Fragment Length Polymorphism
Introduction
Necrotising ulcerative gingivitis (NUG) and nectrotising ulcerative periodontitis (NUP) have been classified as necrotising periodontal diseases (NPD) in the absence of sufficient data that they constitute separate diseases [1]. NPD is a distinct category of periodontal diseases and differs from chronic periodontitis in that it presents as an acute inflammatory condition and patients experience pain and bleeding, with clinically detectable ulceration and necrosis of the interdental papillae and gingival margin [2]. Patients may also present with a fever, bad breath and lymphadenopathy, requiring prompt treatment [3]. While NPD is commonly associated with HIV-infection [4], it is not an uncommon condition among HIV-negative patients, since other systemic disorders and socio-economic factors may play a role [5-7].
Oral candidiasis, with C. albicans as the most commonly associated yeast spp., is an equally well recognized comorbidity of HIV-infection [8,9]. C. albicans colonizes the oral cavity of approximately 50% of healthy individuals [10,11] but is restricted as a commensal organism by a competent host immune response [12-14]. Much interest has also been displayed in the association between yeasts and periodontal disease [15,16].
DNA fingerprinting methods have evolved as reliable tools in the molecular epidemiology of C. albicans [17,18], delineating the organism into 5 major genetic subgroups or clades [17,19,20] and demonstrating a geographic predilection of particular C. albicans clades [19,21,22]. Despite several hundred clinical isolates of C. albicans having been fingerprinted, no evidence emerged of a highly virulent or pathogenic genotype of C. albicans that is associated with disease [17,21,23,24]. C. albicans, propagating in mainly a clonal nature [25], can exist under a wide range of environmental conditions, including co-existing with a variety of other commensal and pathogenic microorganisms through the expression of relevant phenotypic and genotypic characteristics [26-28]. This ability of C. albicans to adapt is also referred to as plasticity [26,29] with the behavior of the organism under conditions of biofilm formation evoking much interest [30-32]. More recently the 'parasexual cycle', with the absence of meiosis, loss of heterozygosity and "extensive karyotypic rearrangement" was suggested as yet another survival mechanism under conditions of environmental stress and it is proposed as a mechanism that may contribute towards genotypic diversity [28,29].
HIV/AIDS and associated mortality and morbidity place a significant demand on scarce health care resources in developing countries where the prevalence of the disease is high [33]. While several previous studies have focused on genotyping of C. albicans isolates obtained from patients presenting with chronic periodontal disease [34-37], uncertainty remains whether antifungal treatment should routinely be included in the treatment of acute periodontal conditions as there is paucity of information on a pathogenic role of C. albicans in such conditions [38]. The absence of fingerprinting studies on C. albicans from NPD prompted this investigation. The objectives of the study were to determine the prevalence of C. albicans in lesions of NPD and to compare that to the prevalence with which the organism occurs on the tongue of the same patient, as well as establishing whether a particularly prevalent genetic subtype of C. albicans is associated with NPD lesions.
Materials and Methods
Patients
The study was approved by the Research, Ethics and Publications Committee of the former University of Limpopo, while informed consent was obtained from the 87 patients included in this study and who presented to the MEDUNSA Oral Health Centre with NPD. Individuals who received antifungal or antibiotic treatment in the 3 months preceding the first visit were excluded from the study, while nevertheless treated for the presenting condition. Oral examinations and treatment were conducted by a single oral medicine-trained examiner using established, presumptive, clinical diagnostic criteria for candidiasis and NPD [1,4]. The extent and severity of NPD was recorded as described by Robinson, et al. [39], with the extent of the NPD lesions categorized as 'few' when ≤ 4 tooth sites were involved and 'many' when ≥ 5 tooth sites were involved, while severity was categorized as 'mild' with tissue necrosis extending ≤ 3 mm from the gingival margin, and 'severe' when tissue necrosis extended ≥ 4 mm from the gingival margin. Treatment of NPD was administered according to an established protocol and patients were urged to return after 5-7 days for a follow-up visit [6]. It was not an objective of the study to establishing a relationship between HIV-status and either the prevalence of C. albicans in lesions of NPD or HIV-status and a particular genetic subtype of C. albicans , but because government policy urges patients to undergo testing [40], the HIV-status of patients is included. All patients of unknown HIV-serostatus were offered HIV-testing with those who agreed to testing providing written consent and receiving pre- and post-test counseling. Based on the test results HIV-positive patients were subsequently referred to a regional HIV-clinic where they were managed according to standard guidelines [41].
Yeast sampling and culture
Before any medication or periodontal treatment was administered, and after the extent and severity of the disease were noted, local anaesthesia was administered and a sterile curette tip was passed along the entire length of the NPD affected sites to collect oral yeasts. During the process the curette tip was frequently shaken in a tube containing 2.5 ml sterile saline before finally dislodged into the tube. A swab was simultaneously taken from the dorsal surface of the tongue by twirling it while stroking across the dorsal surface and also around the border of the tongue. The tube with the curette tip was vortexed at low speed to release any material, the tip removed and the tube again vortexed at full speed for 60 sec to further disrupt the collected material. This was followed by centrifugation at 1 800 × g for 8 min to sediment the collected material. Following this, 2 ml of the supernatant was drawn off and discarded and the sediment was resuspended in the remaining 500 μl, with equal volumes spiral plated on Sabouraud's Dextrose (SD) and CHROMagar plates [42]. The swab taken from the dorsal surface of the tongue was also plated on SD agar and CHROMagar, ensuring contact between the agar and all surfaces of the swab. Plates were incubated at 30 °C for 72 hours. Presumptive identification of C. albicans and other yeasts was performed on colony color on CHROMagar [36,43] and confirmed with the germ tube test. At least 5 colonies presumptively identified as C. albicans were picked from plates that yielded positive yeast growth and stored in sterile water at room temperature until utilised for fingerprinting.
Statistical analyses of the collected data and yeast growth was conducted with SAS®, Release 9, run under Microsoft® Windows® XP for a personal computer. Fisher's Exact Test was performed and p values ≤ 0.05 were considered as significant.
DNA fingerprinting
C. albicans, obtained from both the tongue and NPD of 7 patients, was revived on SD agar.
DNA extraction from these isolates, as well as the reference strain, 3153A, was performed using the method described by Scherer and Stevens [44]. All reagents used were molecular grade, unless otherwise specified. In brief, washed cells were digested with 0.1% β- mercapto-ethanol and 20 μl Zymolyase, before the addition of proteinase K and lysed with 200 μl potassium acetate, purified with phenol:chloroform and DNA in the supernatant was precipitated with 500 μl of 100% isopropanol. RNA was removed with 5 μl of 10 mg/ml RNAase and incubated at 37 °C for 1 hour, followed by a final phenol:chloroform purification and overnight digestion of the extracted genomic DNA at 37 °C with EcoR1 restriction enzyme.
DNA was loaded in the wells of a 0.8% agarose gel and electrophoresed at 65 V over a distance of 16 cm. DNA from the reference strain 3153A was run in the right hand lane. The DNA was transferred from the agarose gel to a nylon membrane through capillary blotting and cross-linked by ultra-violet light. The membrane was hybridized with a 32P-labelled Ca3 probe [45] and radiographed on XO-MAT film, at -20 °C for 24 hrs, before development.
Results
Patients
Demographic and yeast carriage data of 50 (57.5%) females and 37 (42.5%) males are summarised in table 1. Forty patients (46%) declined HIV-testing. Of the 34 confirmed HIV-positive patients, 24 (70.6%) were female, with approximately equal numbers of males and females in the HIV-negative group, as well as the group of unknown HIV-status. Eighty three patients were classified as having extensive lesions (more than 5 teeth involved) and 22 patients classified as severe, with the lesions extending vertically > 3 mm from the gingival margin (Table 2). None of the patients received antiretroviral (ARV) treatment at the time of the first visit and those who tested positive for HIV-infection were referred and managed accordingly at an HIV-clinic of their choice. All of the 35 (40.2%) patients who returned for follow-up, including the 3 patients who were diagnosed with concomitant pseudomembranous candidiasis, presented with complete resolution of their NPD as evidenced by patients reporting absence of pain and bleeding and the absences of gingival ulceration on clinical examination.
![]()
Table 1: Demographic data, HIV-status and isolation of C. albicans from 87 patients presenting with necrotising periodontal disease (NPD).
View Table 1
![]()
Table 2: C. albicans isolated from lesions of NPF in 87 patients in relation to the severity and extent of their NPD.
View Table 2
Yeast culture
C. albicans was cultured from the tongue of 39 (44.8%) patients, while cultured from both the tongue and NPD sites in 7 (8.0%) patients, and from the NPD site but not from the tongue in only one patient (1.1%) (Table 1). Non-C. albicans spp., namely C. krusei and C. parapsilosis, occurred together with C. albicans in two patients and one patient respectively, based on presumptive identification on colony colour on CHROMagar (results not shown).
The relationship between the extent and severity of NPD and yeast carriage among the 87 patients is summarised in table 2. No statistically significant relationship was demonstrated between either the extent (Fisher's Exact Test: p = 0.462) or severity (Fisher's Exact Test: p = 0.3365) and the presence of C. albicans.
DNA fingerprinting
The DNA fingerprinting patterns of the 14 isolates obtained through Southern blot hybridization with the 32P-labeled Ca3 probe, are displayed in figure 1, with the reference strain 3153A in the outermost right hand lane. All the isolates are C. albicans as evident from hybridising with the Ca3 probe. The fingerprinting patterns of 14 (7 pairs) C. albicans isolates revealed genotypic similarity within each pair of isolates from the tongue and from NPD lesion. In assigning a clade affiliation to the pairs the frequency with which bands occur at a particular position, were considered [21]. Isolates 3a and b, and 5a and b displayed a similar dense banding pattern in the 540 and 450 bp region, which is characteristic of the South African clade [21], while the fingerprinting patterns of isolates 2a and b and 4a and b, which are also similar, correspond with that of clade I. Paired isolates 6a and b corresponded to clade II and 7a and b to clade III while 1a and b were not affiliated to any of the known 4 clades that occur among South African oral C. albicans isolates [21,22]. Clinical data and clade affiliation of C. albicans from the 7 patients from whom isolates were obtained from both the tongue and diseased sites, are summarised in table 3.
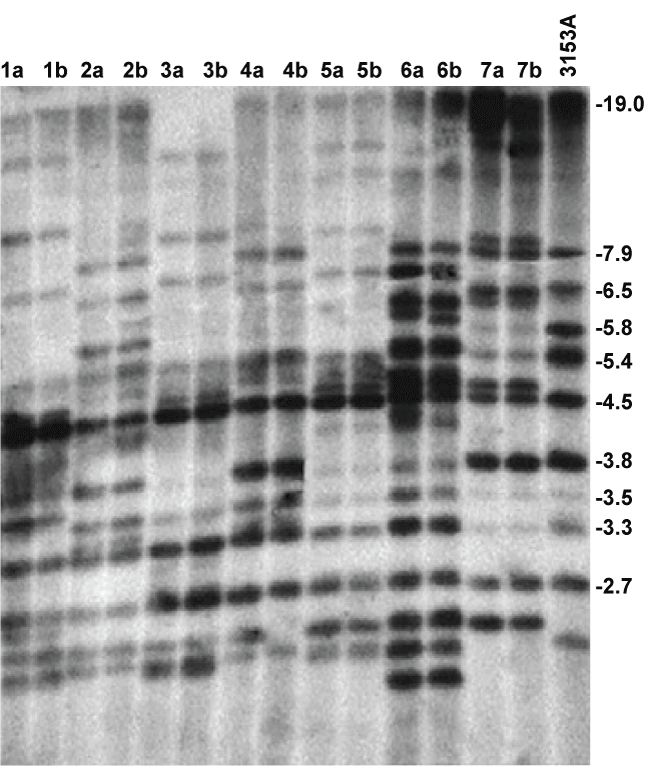
.
Figure 1: Ca3 labeled Southern blot of fingerprinting patterns of 14 isolates of C. albicans obtained simultaneously from NPD lesions and tongue of patients, with 3153A reference strain in the outermost right hand lane.
View Figure 1
![]()
Table 3: HIV-status, clinically diagnosed oral candidiasis, severity and extent of NPD and clade affiliation of paired C. albicans isolates obtained from NPD lesions and the tongue of 7 patients.
View Table 3
Discussion
HIV-infection in developing countries contributes significantly towards morbidity and mortality, placing a high demand on scarce health care resources [33]. Denial and resistance to HIV-testing remains a major stumbling block in curbing the disease [46]. No reliable conclusions can be made regarding HIV-infection and any presenting pathology when approximately half of the patient population is of unknown HIV-status, hence the exclusion thereof from the objectives of this study. The 60% loss to follow-up among this cohort corresponds with elsewhere in South or sub-Saharan Africa [47]. Despite these particular socio-demographic factors, the unnecessary administration of medication cannot be afforded for economic as well as reasons of sound clinical practice. The clinical findings of this study, excluding the 3 patients who initially presented with pseudomembranous candidiasis and received antifungal treatment, as well as previous studies, confirm that NPD resolved without the administration of antifungal agents [5,6,48]. Because both NPD and oral candidiasis are associated with immunosuppressive conditions [9] it is likely that immunosuppressed patients would either carry a higher load of C. albicans on the oral mucosa [49] or that the two disease conditions occurred simultaneously [15,39] and C. albicans being perceived as playing a causative role in NPD.
This was the first fingerprinting, or genotyping study to compare C. albicans from lesions of NPD with those obtained from elsewhere in the mouth, namely the tongue. Restriction fragment length polymorphism (RFLP), combined with the Ca3 labeled probe, is accepted as a reproducible fingerprinting method with high discriminatory power [18,29], as demonstrated by significant numbers of clinical C. albicans isolates from different geographic areas or patient cohorts that were fingerprinted according to this method [22]. In this study clearly discernable fingerprinting patterns were obtained (Figure 1), revealing similar patterns between the two isolates of each pair for all of the 7 pairs. This is strongly suggestive of contamination between the two sites and because the organism had a much lower prevalence in NPD sites that the tongue was the most likely the primary site of colonisation [10]. Assigning a clade affiliation to the fingerprinted isolates was readily achieved due to the similarity within the pairs and the high degree of resolution that the Southern blot (Figure 1) provided. The representation of the different clades in this study corresponded with the findings of a previous study on South African oral C. albicans isolates in which the majority of isolates from both HIV-positive and healthy subjects clustered in clade SA and clade I [21]. Taking the prevailing clade distribution of C. albicans in a particular geographic locale into consideration is required in not mistaking isolates from particular anatomical sites or disease conditions as being 'unique' or 'enriched'.
The authors are confident that the 9.2% prevalence of yeasts from NPD sites is not an underestimation as specimens were collected along the entire affected gingival margin that involved more than 5 teeth in 83 of the 87 patients. The prevalence obtained in this study corresponds well with the 10%, 13% and 15,6% reported respectively by Moore, et al. [50], Lamster, et al. [35] and Reynaud, et al. [51]. As it is not possible to standardise the collection of yeast specimens when using a curette as opposed to paper points, no attempt was made to quantify or make deductions on the number of colony forming units (cfu) obtained from the various patients or the two sites. The oral rinse method cannot be used when comparing the prevalence with which yeasts can be isolated from two different oral sites, as in this study, due to the swishing action resulting in an 'artificial' spread of organism throughout the mouth. To have fingerprinted several C. albicans isolates from the same patient is unlikely to have changed the findings as several studies have proven that the same genotype of the organism was repeatedly isolated from the same individual [17,18], with possible microevolutionary changes in the hypervariable region of the genome [52]. It is therefore unlikely that the genotypic similarity of isolates from two sites in all 7 patients can be attributed to chance. Alternatively, if it was common that patients are colonised by a variety of distinctly different genetic subtypes of C. albicans, it would be unlikely that the two isolates, one obtained from the tongue and the other from NPD, would be identical. Fourteen fingerprinted isolates therefore constitute a sufficient number to make deductions from. In addition, a most recent study, using the same fingerprinting method as in this study, demonstrated the colonisation by a similar genetic subtype of C. albicans from oral mucosa and carious dentine [53]. The lack of association between C. albicans and NPD is further substantiated by the fact that no statistically significant association between either the severity or the extent of NPD and the carriage of oral yeasts among this cohort could be established (Table 2). The 52.8% of patients from whom isolates were obtained from the tongue also fall within the normal range of oral Candida carriage reported for both healthy and HIV-positive patients without clinically detectable candidiasis [10,11,37].
Previous fingerprinting studies on C. albicans from sites of chronic periodontitis, healthy gingival sulci and, or oral mucosa, all used different fingerprinting methods [34-37,54]. Three of the studies reported unique C. albicans genotypes isolated from sub-gingival areas or periodontal pockets [35,36,55], one not finding any difference between genotypes and the different oral site they were isolated from [34] and yet another reporting an "enrichment" of a particular clade [54]. The latter finding might well be due to the natural clade distribution of C. albicans clades in that geographic area [17], while the other studies did not explain their findings apart from mentioning that these unique strains did not originate from elsewhere in the mouth [35]. These studies on C. albicans from chronic periodontitis sites were all performed before the most recent discoveries on how C. albicans adapts to harsh environmental conditions and in the process generates genomic variance [28]. Considering the conditions in the mouth, it is attractive to speculate that genomic variance in C. albicans may have actually resulted in sites, or pockets of chronic periodontal disease. In periodontal pockets deeper than 3 mm there is low oxygen tension (anaerobiosis), oral biofilm is protected from disruption, allowing it to mature [56,57] while there is interaction between C. albicans and other oral microflora, including host defenses [28,29,58]. It is proposed that oral biofilm in periodontal pockets provide a good in vivo niche conducive to C. albicans mating and for the parasexual cycle to produce "tetraploid progenitors (which) can produce populations of progeny cells with a high degree of genomic diversity, …" [58]. Future in vivo studies on oral biofilm, for example sub- or supragingival plaque in healthy gingival tissue or chronic periodontal disease, pseudomembranous or chronic hyper plastic candidiasis, could offer an opportunity to further elucidate findings from laboratory or mouse model studies on the occurrence of 'unique' genetic subtypes [28,58]. Longitudinal observations on isolates from a state of disease through to resolution thereof, may provide valuable answers regarding the "fitness" of C. albicans strains that have possibly undergone genotypic rearrangement, to survive in the human host, as well as offering information on a possible shift in the expression of relevant phenotypic characteristic at each point in time [17,28,31,57,58].
In conclusion it is emphasized that this is the first study in which C. albicans isolates from acute periodontal disease were fingerprinted and that the results differ from that found in chronic periodontitis or healthy tissue. It is possible that the dynamics of the acute inflammatory environment, with an abundance of necrotic, desquamating tissue, associated bleeding and mediators of acute inflammation [2,3], is not conducive to the undisturbed colonization, replication and biofilm formation by C. albicans. The low frequency with which C. albicans was isolated from sites of NPD, the absence of a statistically significant correlation between NPD and C. albicans carriage, the condition resolving without antifungal treatment, as well as the genotypic similarity between paired isolates from diseased sites and the tongue contribute to the conclusion that it is unlikely that highly virulent strains of C. albicans exist. No such strain or strains were demonstrated to have played a role or contributed to the pathogenesis of NPD in this instance. Furthermore, for the first time, recent sophisticated in vitro research findings on the parasexual cycle of C. albicans is discussed in a clinical context, namely chronic periodontal disease.
Funding Information
South African Medical Research Council (MRC) grant to EB. The funders had no role in the design of the study, data collection and interpretation, or the decision to submit the work for publication. There are also no other conflicts of interest.
Acknowledgement
Laboratory assistance from Me. E. Sekati and J. Molepo.
References
-
Armitage GC (1999) Development of a classification system for periodontal diseases and conditions. Ann Periodontol 4: 1-6.
-
Herrera D, Alonso B, de Arriba L, Santa Cruz I, Serrano C, et al. (2014) Acute periodontal lesions. Periodontol 2000 65: 149-177.
-
Albandar JM (2014) Aggressive and acute periodontal diseases. Periodontol 2000 65: 7-12.
-
Shiboski CH, Patton LL, Webster-Cyriaque JY, Greenspan D, Traboulsi RS, et al. (2009) The Oral HIV/AIDS Research Alliance: updated case definitions of oral disease endpoints. J Oral Pathol Med 38: 481-488.
-
Phiri R, Feller L, Blignaut E (2010) The severity, extent and recurrence of necrotizing periodontal disease in relation to HIV status and CD4+ T cell count. J Int Acad Periodontol 12: 98-103.
-
Shangase L, Feller L, Blignaut E (2004) Necrotising ulcerative gingivitis/periodontitis as indicators of HIV-infection. SADJ 59: 105-108.
-
Wood NH, Blignaut E, Lemmer J, Meyerov R, Feller L (2011) Necrotizing periodontal diseases in a semirural district of South Africa. AIDS Res Treat 2011: 638584.
-
Arendorf T, Holmes H (2000) Oral manifestations associated with human immunodeficiency virus (HIV) infection in developing countries--are there differences from developed countries? Oral Dis 6: 133-135.
-
Greenspan JS (1997) Sentinels and signposts: the epidemiology and significance of the oral manifestations of HIV disease. Oral Dis 1: S13-17.
-
Arendorf TM, Walker DM (1980) The prevalence and intra-oral distribution of Candida albicans in man. Arch Oral Biol 25: 1-10.
-
Odds F (1988) Candida and Candidosis: a Review and Bibliography. (2nd edn), Bailliere Tindall, London.
-
Cheng SC, van de Veerdonk F, Smeekens S, Joosten LA, van der Meer JW, et al. (2010) Candida albicans dampens host defense by downregulating IL-17 production. J Immunol 185: 2450-2457.
-
Conti HR, Gaffen SL (2010) Host responses to Candida albicans: Th17 cells and mucosal candidiasis. Microbes Infect 12: 518-527.
-
Naglik JR, Richardson JP, Moyes DL (2014) Candida albicans pathogenicity and epithelial immunity. PLoS Pathog 10: e1004257.
-
Cobb CM, Ferguson BL, Keselyak NT, Holt LA, MacNeill SR, et al. (2003) A TEM/SEM study of the microbial plaque overlying the necrotic gingival papillae of HIV-seropositive, necrotizing ulcerative periodontitis. J Periodontal Res 38: 147-155.
-
Jabra-Rizk MA, Ferreira SM, Sabet M, Falkler WA, Merz WG, et al. (2001) Recovery of Candida dubliniensis and other yeasts from human immunodeficiency virus-associated periodontal lesions. J Clin Microbiol 39: 4520-4522.
-
Odds FC (2010) Molecular phylogenetics and epidemiology of Candida albicans. Future Microbiol 5: 67-79.
-
Soll DR (2000) The ins and outs of DNA fingerprinting the infectious fungi. Clin Microbiol Rev 13: 332-370.
-
Bougnoux ME, Aanensen DM, Morand S, Theraud M, Spratt BG, et al. (2004) Multilocus sequence typing of Candida albicans: strategies, data exchange and applications. Infect Genet Evol 4: 243-252.
-
Chowdhary A, Lee-Yang W, Lasker BA, Brandt ME, Warnock DW, et al. (2006) Comparison of multilocus sequence typing and Ca3 fingerprinting for molecular subtyping epidemiologically-related clinical isolates of Candida albicans. Med Mycol 44: 405-417.
-
Blignaut E, Pujol C, Lockhart S, Joly S, Soll DR (2002) Ca3 fingerprinting of Candida albicans isolates from human immunodeficiency virus-positive and healthy individuals reveals a new clade in South Africa. J Clin Microbiol 40: 826-836.
-
Soll DR, Pujol C (2003) Candida albicans clades. FEMS Immunol Med Microbiol 39: 1-7.
-
L'Ollivier C, Labruère C, Jebrane A, Bougnoux ME, d'Enfert C, et al. (2012) Using a Multi-Locus Microsatellite Typing method improved phylogenetic distribution of Candida albicans isolates but failed to demonstrate association of some genotype with the commensal or clinical origin of the isolates. Infect Genet Evol 12: 1949-1957.
-
Bougnoux ME, Diogo D, Francois N, Sendid B, Veirmeire S, et al. (2006) Multilocus sequence typing reveals intrafamilial transmission and microevolutions of Candida albicans isolates from the human digestive tract. J Clin Microbiol 44: 1810-1820.
-
Bougnoux ME, Pujol C, Diogo D, Bouchier C, Soll DR, et al. (2008) Mating is rare within as well as between clades of the human pathogen Candida albicans. Fungal Genet Biol 45: 221-231.
-
Soll DR (2002) Candida commensalism and virulence: the evolution of phenotypic plasticity. Acta Trop 81: 101-110.
-
Krom BP, Kidwai S, Ten Cate JM (2014) Candida and other fungal species: forgotten players of healthy oral microbiota. J Dent Res 93: 445-451.
-
Forche A (2014) Large-Scale Chromosomal Changes and Associated Fitness Consequences in Pathogenic Fungi. Curr Fungal Infect Rep 8: 163-170.
-
McManus BA, Coleman DC (2014) Molecular epidemiology, phylogeny and evolution of Candida albicans. Infect Genet Evol 21: 166-178.
-
Daniels KJ, Park YN, Srikantha T, Pujol C, Soll DR (2013) Impact of environmental conditions on the form and function of Candida albicans biofilms. Eukaryot Cell 12: 1389-1402.
-
Daniels KJ, Srikantha T, Pujol C, Park YN, Soll DR (2015) Role of Tec1 in the development, architecture, and integrity of sexual biofilms of Candida albicans. Eukaryot Cell 14: 228-240.
-
Soll DR (2011) Evolution of a new signal transduction pathway in Candida albicans. Trends Microbiol 19: 8-13.
-
UNAIDS (2015) Country Reports.
-
Barros LM, Boriollo MF, Alves AC, Klein MI, Goncalves RB, et al. (2008) Genetic diversity and exoenzyme activities of Candida albicans and Candida dubliniensis isolated from the oral cavity of Brazilian periodontal patients. Arch Oral Biol 53: 1172-1178.
-
Lamster IB, Grbic JT, Mitchell-Lewis DA, Begg MD, Mitchell A (1998) New concepts regarding the pathogenesis of periodontal disease in HIV infection. Ann Periodontol 3: 62-75.
-
Pizzo G, Barchiesi F, Falconi Di Francesco L, Giuliana G, Arzeni D, et al. (2002) Genotyping and antifungal susceptibility of human subgingival Candida albicans isolates. Arch Oral Biol 47: 189-196.
-
Song X, Eribe ER, Sun J, Hansen BF, Olsen I (2005) Genetic relatedness of oral yeasts within and between patients with marginal periodontitis and subjects with oral health. J Periodontal Res 40: 446-452.
-
Robinson PG (1997) Treatment of HIV-associated periodontal diseases. Oral Dis 3 Suppl 1: S238-240.
-
Robinson PG, Sheiham A, Challacombe SJ, Wren MW, Zakrzewska JM (1998) Gingival ulceration in HIV infection. A case series and case control study. J Clin Periodontol 25: 260-267.
-
SANAC (2010) The National HIV Counselling and Testing Campaign Strategy.
-
SANDH (2015) National Consolidated Guidelines.
-
Odds FC, Bernaerts R (1994) CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J Clin Microbiol 32: 1923-1929.
-
Beighton D, Ludford R, Clark DT, Brailsford SR, Pankhurst CL, et al. (1995) Use of CHROMagar Candida medium for isolation of yeasts from dental samples. J Clin Microbiol 33: 3025-3027.
-
Scherer S, Stevens DA (1987) Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol 25: 675-679.
-
Anderson J, Srikantha T, Morrow B, Miyasaki SH, White TC, et al. (1993) Characterization and partial nucleotide sequence of the DNA fingerprinting probe Ca3 of Candida albicans. J Clin Microbiol 31: 1472-1480.
-
Zuma K, Shisana O, Rehle TM, Simbayi LC, Jooste S, et al. (2016) New insights into HIV epidemic in South Africa: key findings from the National HIV Prevalence, Incidence and Behaviour Survey, 2012. Afr J AIDS Res 15: 67-75.
-
Losina E, Bassett IV, Giddy J, Chetty S, Regan S, et al. (2010) The "ART" of linkage: pre-treatment loss to care after HIV diagnosis at two PEPFAR sites in Durban, South Africa. PLoS One 5: e9538.
-
Feller L, Buskin A, Blignaut E (2005) A review of candida and periodontal disease in immunocompetent and HIV-infected subjects. SADJ 60: 152-154.
-
Alt-Epping B, Nejad RK, Jung K, Gross U, Nauck F (2012) Symptoms of the oral cavity and their association with local microbiological and clinical findings--a prospective survey in palliative care. Support Care Cancer 20: 531-537.
-
Moore LV, Moore WE, Riley C, Brooks CN, Burmeister JA, et al. (1993) Periodontal microflora of HIV positive subjects with gingivitis or adult periodontitis. J Periodontol 64: 48-56.
-
Reynaud-Mendel B, Janier M, Gerbaka J, Hakim C, Rabian C, et al. (1996) Dermatologic findings in HIV-1-infected patients: a prospective study with emphasis on CD4+ cell count. Dermatology 192: 325-328.
-
Pujol C, Joly S, Nolan B, Srikantha T, Soll DR (1999) Microevolutionary changes in Candida albicans identified by the complex Ca3 fingerprinting probe involve insertions and deletions of the full-length repetitive sequence RPS at specific genomic sites. Microbiology 145: 2635-2646.
-
Blignaut E, van Heerden WF (2015) Molecular and Histological Association Between Candida albicans from Oral Soft Tissue and Carious Dentine of HIV-Positive Children. Mycopathologia 180: 193-201.
-
McManus BA, Maguire R, Cashin PJ, Claffey N, Flint S, et al. (2012) Enrichment of multilocus sequence typing clade 1 with oral Candida albicans isolates in patients with untreated periodontitis. J Clin Microbiol 50: 3335-3344.
-
Song X, Sun J, Støre G, Hansen BF, Olsen I (2009) Colony morphologies, species, and biotypes of yeasts from thrush and denture stomatitis. Acta Odontol Scand 67: 248-255.
-
Soll DR (2009) Why does Candida albicans switch? FEMS Yeast Res 9: 973-989.
-
Soll DR (2014) The role of phenotypic switching in the basic biology and pathogenesis of Candida albicans. J Oral Microbiol 6.
-
Hickman MA, Paulson C, Dudley A, Berman J (2015) Parasexual Ploidy Reduction Drives Population Heterogeneity Through Random and Transient Aneuploidy in Candida albicans. Genetics 200: 781-794.





