The genus Aeromonas is widely distributed in aquatic environments and certain strains are serious pathogens in poikilothermic animals and humans. Human infections related with this pathogen include gastroenteritis, urinary tract infections, pneumonia, wound infections and septicemia. In this study, both the correct medical diagnosis with taking advantages of molecular approaches and a brief investigation of the enzymatic activity potential of Aeromonas were aimed. Aeromonas spp. isolates were collected from different stream samples of Sinop, Turkey and were identified according to morphological, physiological and biochemical tests based on API 20NE (Biomeriux, France). The evaluation of total cell protein profiles and molecular identification based on 16S rRNA sequencing were conducted. Amylase, cellulase and protease enzyme activities of strains were also determined. The strains were separated into two clusters of 73% and over according to the SDS-PAGE total cell protein profiles reflecting a less discriminative power for this method in the strain identification. On the other hand, according to the 16S rRNA sequence analysis results, two isolates were identified as Aeromonas tecta and the other two as Aeromonas veronii at 99% similarity level reflecting the superiority of this approach when compared with SDS-PAGE profile. Enzyme assays showed that; three of the isolates were able to produce the amylase and only one could produce the cellulase but none produced the protease enzyme. Though our results are based on limited samples, enzyme activity potential of Aeromonas is recommended to be further investigated in detail in future studies. Pollution of the water by organic materials is a serious environment and health problem and thus bacterial extracellular enzyme activities degrading these pollutants in aquatic environments may reflect a bioremediation strategy in terms of a biotechnological perspective. Taking into consideration of Aeromonas in human infections, the realiable identification based on molecular techniques is an obligatory for medical practice. Besides, exploiting this pathogen in search of enzyme activities may broaden our biotechnology perspective.
Aeromonas, SDS-PAGE, Molecular characterization, 16S rRNA, Protease, Amylase, Cellulase
Aeromonas species are Gram-negative, rod-shaped facultatively anaerobic bacteria and ubiquitous in aquatic environments, different types of food, also in invertebrate and vertebrate organisms [1]. Certain Aeromonas strains are serious pathogens in poikilothermic animals, birds and mammals. Human infections include gastroenteritis, urinary tract infections, pneumonia, wound infections and septicemia [2,3]. Potential pathogenicity of Aeromonas spp. stems from the expression of several virulence factors including inherent resistance to various antibiotics and ability to form biofilms using quorum sensing [1,2]. Though the effect of Aeromonas in medicine is clear enough, the biotechnological processes such as biodegradation of toxic organic pollutants and thus developing bioremediation strategies it can be involved in is a field needs to be further investigated. This study aimed to propose both the medical diagnosis with the comparison of molecular approaches and potential biotechnological role of Aeromonas with the aim of enrichment in future comprehensive studies.
Conventional biochemical tests are important for biotyping Aeromonas isolates however they lack of discriminatory power to correctly identify at the species level and thus the use of various genotyping methods are inevitable. In this study, we aimed to correctly characterize this waterborne opportunistic pathogen with SDS-PAGE total cell protein profiles and 16S rRNA sequencing approaches. In addition, some enzymatic activities of the isolates were determined for the future utilization potential of these pathogens in the biotechnology area. Our preliminary results can direct future researchers to conduct comprehensive studies required in this field.
A total of 27 samples from surface stream sampling sites were collected over a one year period (September to November, 2016) in Sinop, Turkey and analyzed for the presence of Aeromonas species based on API 20NE tests for the identification and speciation of Aeromonas. Sampling was carried out at a depth of 15 cm. Water samples were collected in 100 ml autoclaved sterile bottles and immediately placed on ice in coolers, and delivered to the laboratory. The samples were processed within 3 h of collection time. The isolates were grown on Nutrient agar at 37 ℃ for 18-24 hours. Four Aeromonas isolates were identified from surface stream sampling sites in Sinop, Turkey.
The strains were identified as Aeromonas according to API 20NE (Biomeriux, France) identification kit, Gram staining and reproductive abilities at physiological conditions such as different temperature, salinity and pH.
Extraction of whole cell proteins was carried out according to the method of Laemmli [4]. Protein analysis was carried out with SDS-PAGE in gel slabs of 1 mm thickness (4% stacking and 10% resolving gels for total proteins). The molecular weight of each band was calculated with Total Lab 1D Manual R11.1, UK programme.
Chromosomal DNA extraction from Aeromonas strains was carried out according to the method of Sambrook, et al. [5]. DNA samples were stored at -20 ℃ until further molecular analysis. Sequence analysis of the 16S rRNA gene regions was performed for genotypic tests with primer pairs 519F and 1492R [6]. Primer sequences are shown in Table 1. The following PCR conditions were applied to each assay; 0.75 mM MgCl2, 100 µM dNTPs, 25 pmol of forward ad reverse primers, 5 U Taq DNA polymerase (Thermo Fisher Scientific), 1 µl DNA per 50 µl reaction. The thermal parameters at Techne TC-5000 (California-USA) were as follows: Initial denaturation at 95 ℃ for 7 min, followed by 25 cycles of denaturation at 94 ℃ for 1 min, annealing at 54 ℃ for 1 min and extension at 72 ℃ for 90 sec. A final extension at 72 ℃ for 6 min at the end of the cycles was included. The amplicons obtained from 16S rRNA region were resolved by 1.8% agarose gel electrophoresis stained with ethidium bromide and visualized under UV transilluminator (Cleaver-MicroDOC, UK). The PCR products were purified and sent to sequencing analysis to MedSanTek biotechnology firm. Sequences generated from different isolates were compared to the GenBank database using the BLAST (Basic Local Alignment Search Tool) maintained by the National Center for Biotechnology Information and multiple sequence alignments were made with the Clustal W. The alignment was further analyzed using MEGA.7.0 (Molecular Evolutionary Genetics Analysis Version 7.0) program with Neighbour-Joining (NJ) method and phylogenetic tree was constructed.
Table 1: Primer sequences used in the study. View Table 1
Enzyme production activities of isolates in Skim milk agar (protease), M9 agar (amylase) and CMC agar (cellulase) were performed at 25, 35 and 45 ℃ at different pH (5-10).
The isolated strains were streaked on pH: 7 skim milk agar and incubated at 37 ℃ for 24 h. Proteolytic bacteria use the enzyme caseinase to hydrolyze casein and form soluble nitrogenous compounds displayed as a clear zone around colonies. The nonappearance of a clear zone around colonies was evaluated as protease negative.
The isolated Aeromonas strains were streaked on pH-7 M9 starch agar plate and incubated at 37 ℃ for 72 h. Following incubation, the plates were flooded by iodine to produce starch-iodine complex which is visualized by deep blue color. The appearance of clear zone at starch agar plate was used as the main indicator of amylase positivity.
The isolated strains were streaked on pH-7 CMC agar plate and incubated at 37 ℃ for 24 h. At the end of the incubation period, the strains were stained with iodine solution for 5 min. The appearance of clear zone around colonies was evaluated as positive.
All the four isolates were able to grown in 2% NaCl at pH values 6-7 and they were positive for katalase and Voges-Proskauer reaction. All the isolates were grown at temperatures 25 ℃ and 37 ℃, only one isolate (Z1) were grown at 45 ℃, and no growth was observed at 55 ℃ and over. According to the fermentation analysis results, all the four isolates were positive for sucrose, D-mannose, maltose, D-glucose, D-manitol, and D-xylose and negative for D-sorbitol.
Biochemical, temperature, and fermentation test results of Aeromonas isolates are shown in Tables 2, Table 3, and Table 4.
Table 2: Biochemical analysis. View Table 2
Table 3: Temperature analysis. View Table 3
Table 4: Fermentation analysis. View Table 4
According to SDS-PAGE total cell protein profiles, the strains were separated into two major clusters between 73% and over (Figure 1). First cluster showed 78% similarity level and second cluster showed 81% similarity level. Protein electrophoretic patterns of the isolates yielded between 22-26 bands. Protein profiles were very similar among the strains and several strains exhibited dominant protein bands.
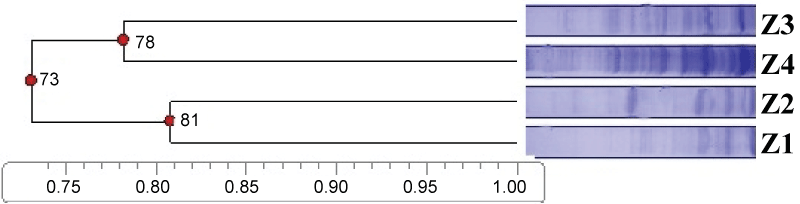 Figure 1: Dendrogram derived from cluster analysis (UPGMA) illustrating SDS-PAGE analysis of whole cell proteins of Aeromonas isolates.
View Figure 1
Figure 1: Dendrogram derived from cluster analysis (UPGMA) illustrating SDS-PAGE analysis of whole cell proteins of Aeromonas isolates.
View Figure 1
Sequence analysis of the 16S rRNA gene regions was performed for genotypic tests and compared to the sequences recorded in GenBank. GenBank accession numbers for the isolates were also determined: MH444890, MH444891, MH444892, and MH444893. Homology search carried out using BLAST (http://www.ncbi.nlm.nih.gov) revealed that the two isolates were similar to Aeromonas tecta and the other two were close to Aeromonas veronii at 99% similarity level. When the alignment was further analyzed using MEGA.7.0 Neighbour-Joining (NJ) method, the phylogenetic tree constructed reflected the evolutionary relationships of the isolates (Z1-Z2-Z3-Z4) with the other strains in the genus (Figure 2).
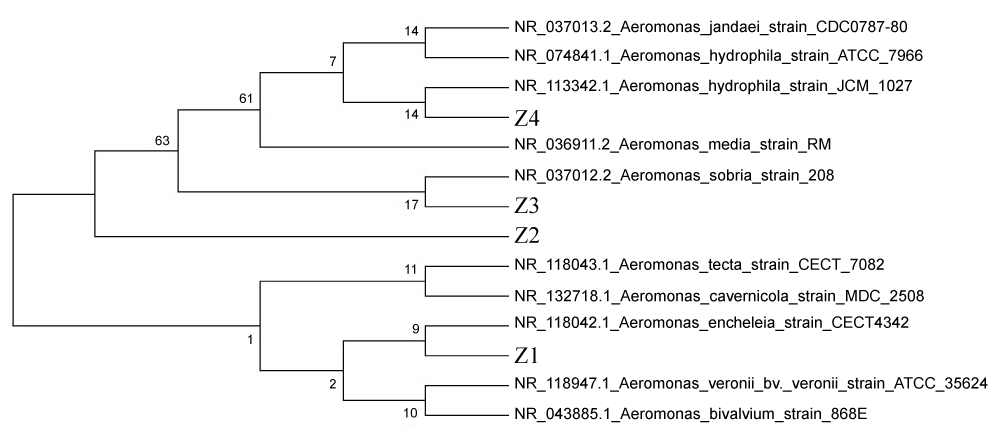 Figure 2: Evolutionary history of the taxa analyzed.
View Figure 2
Figure 2: Evolutionary history of the taxa analyzed.
View Figure 2
According to results obtained, three of the isolates were able to produce the amylase enzyme and only one could produce the cellulose enzyme but none produced the protease enzyme. For the amylase enzyme, in particular at 25 ℃, one strain formed a 12-16 mm zone diameter in the pH range of 6-10, the second strain for 35 ℃ formed a 13 mm zone diameter at pH-6, and for 45 ℃ only the other two strains in the pH (6-10) had a low zone diameter of 2-4 mm. The cellulose enzyme producing strain was found to have zone diameters of 23 and 13 mm at pH-7 for 25 ℃ and 45 ℃, respectively. Results of amylase and cellulose production at different temperature and pH values are depicted in Tables 5 and Table 6.
Table 5: Amylase Production Analysis. View Table 5
Table 6: Cellulase Production Analysis. View Table 6
Since Aeromonas is widely distributed in aquatic environments, serious cautions must be taken. Fraisse, et al., presented a retrospective study of Aeromonas spp. infections between years 1997-2004 and stated that ten infections were nosocomial and four were associated with medical leeches [7]. Voss, et al. stated that 13 of 28 Aeromonas wound and soft tissue infections during a 4-year-period were associated with a water-related injury and lakes or rivers constituted 43% of the total [8]. Extracellular proteases allow Aeromonas sp. to persist in different environments and thus facilitate ecological interactions with other organisms and promote invasion by direct damage of host tissue [3]. Therefore, the investigated enzymes such as proteases are not only important in terms of biotechnological perspective, but also they can be a measure of improvement of survival and infection potential in aquatic environments. Since none of our isolates produced the protease enzyme, this can also be a measure of low infectivity potential in the aquatic areas investigated.
Using various conditions for the biochemical identification of Aeromonas spp. is one of the major problems that gives unstable results in clinical laboratories. Thus, realiable method choice to correctly identify Aeromonas species can help to prevent the inconsistence for this complicated taxonomic genus. Though API strips have database for the genus Aeromonas, they are effective in the identification at the genus level since they lack the database for the entire range of the known Aeromonas species [9]. The correct method approaches will be beneficial to clarify the identification of Aeromonas spp. at the species level and support routine biochemical identification in laboratories. One of these approaches can be supporting the routine phenotyping tests with multiplex PCR though it also has some limits in final discrimination and therefore must not be used as a single identification approach [1]. Sarkar, et al., also implied the less discriminatory power of SDS-PAGE and offered Random Amplied Polymorphic DNA (RAPD) analysis as an ideal tool for molecular characterization [10]. Demarta, et al., characterized clinical and environmental Aeromonas isolates by a polyphasic approach including phylogenetic analysis derived from gyrB, rpoD, and 16S rRNA gene sequencing as well as DNA-DNA hybridization together with extensive biochemical tests and described a novel species named as Aeromonas tecta [11].
The less discriminative power of SDS-PAGE at strain level demands more specialized molecular tools alone or in combination and sequence analysis of the 16S rRNA gene regions corresponds to this requirement. The combination of phenotyping and a strong molecular approach ensures the accurate identification of Aeromonas spp. in medical laboratories.
Aeromonas hydrophila is the predominant species in freshwater and municipal drinking water supplies and in Nigeria lakes, the isolated species were A. hydrophila and A. sobria [12]. Several Aeromonas species having potential pathogenicity were also investigated in environmental water of Okinawa and A. veronii biovar sobria and A. jandaei were reported as the predominant species in dams, and A. hydrophila and/or A. eucrenophila were predominant in wells [13]. However, our limited number of isolates did not allow us to characterize the predominant species A. hydrophila and extended studies are certainly required.
Enzyme production activities of the isolates in Skimmilk agar (protease), M9 agar (amylase) and CMC agar (cellulase) were also determined at 25, 35 and 45 ℃ at different pH (5-10). Besides with other various industrial applications of microbial enzymes, utilizing these microbial enzyme systems may also be involved in the biodegradation of toxic organic pollutants. Soil and water pollution by industrial chemicals and petroleum hydrocarbons is a serious modern world problem and the use of bioremediation strategies may offer nature friendly approach to remove/reduce these contaminants [14,15]. Our results showed that three of the isolates were able to produce the amylase enzyme and one could produce the cellulase enzyme. However, none produced the protease enzyme. Though this is a preliminary analysis with limited number, our results reflect the potential of Aeromonas in biotechnological field besides with the widely studied bacteria and fungi. We encourage future comprehensive studies focusing on this perspective.
Since Aeromonas species are opportunistic waterborne pathogens and can cause health problems, continuous surveillance of water sources is an important issue to protect susceptible individuals. Also, the realiable identification methods based on molecular techniques is an obligatory in medical practice for this phenotypically and genetically heterogeneous taxon. Besides, exploiting this pathogen in search of enzyme activities may have area of utilization in developing bioremediation strategies.
We thank to Prof. Dr. Sumer ARAS.