Having been identified for centuries, breast cancer still remains one of the leading causes of death in women. Within the United States of America (USA) alone, it is estimated that over 60,000 women would have developed new cases of malignant breast cancer in 2017 alone; thus, calling an attention for the development of a reliable and effective breast cancer model for drug discovery to combat the disease or diagnostics to predict the progression of the disease. Current breast cancer models involve usage of 2D cell cultures and in vivo models which do not recapitulate the entire microenvironment of tumour growth in human cases, and despite some cell lines being derived from patients' own tumour cells, 2D cell cultures are heterogeneous and lack the cell-cell interaction necessary for normal tumour growth in the human body. Many limitations of these conventional models can be resolved using novel 3D organoid cultures produced from tumours of patients themselves or other viable sources. Breast cancer organoids have been proven to predict patient chemotherapeutic outcomes efficiently in the case of primary breast cancer via immunofluorescence or gene expression studies. This study model can also be utilized to discover new biomarkers with clinical implications. The advantages, limitations, and protocols to derive breast cancer organoids are being reviewed here alongside the ethical issues which might arise. Breast cancer organoid research could then open avenues of reliable drug discovery and diagnostics model to ameliorate subtypes of breast cancer which do not provide much options therapy-wise, such as the triple negative subtype.
3D cell culture, Breast cancer, Organoid, Patient-derived xenografts, Spheroid
The study of breast cancer has progressed massively over the past 2 decades, which raised public awareness of this disease as well as the quality of treatment. Despite the massive amount of effort, the effectiveness of clinically breast cancer chemotherapeutic drugs, including tamoxifen and anastrozole [1,2], as well as trastuzumab [3], a widely known drug which specifically targets human epidermal growth factor 2 (HER2) breast cancer, still varies from one patient to another [3,4]. Having understood how similarly but also differently the molecular progression of different patients' cases calls for the attention to develop a functional tailored biological model, with the goal of effectively curing this disease, one patient at a time.
Throughout the years, various breast cancer cell lines have been established including the widely used Michigan Cancer Foundation-7 (MCF-7) [5] and Monroe Dunaway Anderson (MDA-MB-231) [6,7] invasive ductal carcinoma cell lines amongst others. Despite the major breakthroughs these cancer cell lines in the drug discovery or understanding the basic molecular mechanism of disease progression, it has become apparent that 2D cultures of breast cancer is not enough to fully conclude the efficacies of different treatments in patients, especially in a dynamic disease which incorporate intercalating components [8,9]. Moreover, the use of animal models (e.g. rodents, cats, and dogs) [10] in the study of human breast cancer, albeit some physiological matches, still does not portray the disease as a whole [11,12]; thus, as medical sciences progress to more personalized variants of treatments, diagnostic and predictive models of breast cancer should also provide a more specific and holistic approach.
Turning away from conventional 2D cell lines, researchers started cultivation of 3D cell cultures, commonly known as spheroids, popularized by the various works of Bissell, et al. which underlined the importance of tumour microenvironment which can be recapitulated by a more robust 3D cell culture [13]. Producible within hydrogel scaffolds [14,15] or bioreactors [16,17], these spheroids can be further developed to form the more in vivo-like organoids which express most components displayed by cancer cells, including malignant epithelial and endothelial cells aside from extracellular matrix (ECM) components [12,18]. This article will then review the rapidly emerging organoid models in breast cancer research alongside its advantages and limitations in the face of currently available 2D or animal models.
Breast cancer is one of the first cancers to be researched vastly and the heterogeneous nature of this disease has been observed since before the dawn of molecular studies. Early histopathological findings have provided researchers with different physiological insights, including distinctions between ductal and lobular traits or the infiltration of metastases into the lymph nodes [19,20]. Currently, there are several molecular subtypes of breast cancer, all of which pose different clinical implications and outcome based on the course of treatment the patient undergoes. Based on the breakthrough provided by Sørlie, et al. with their microarray studies of ample complementary deoxyribonucleic acid (cDNA) from breast cancer samples, 5 molecular subtypes of breast cancer could be deduced [21]. These molecular subtypes differ in the grades of the tumour and presence (i.e. positive or negative) of oestrogen receptor (ER), progesterone receptor (PR), HER2, and Ki67 protein [21,22]. The status within each subtype is summarized in Figure 1 constructed below. Other studies yielded similar molecular classifications derived their grouping system via microarray analysis, such classifications were made in studies conducted by Munirah, et al. which discovered that majority of Malay ethnic breast cancer cases belong to the luminal A subtype, and unlike other subtypes in this classification, it is accompanied by negativity of the p53 protein [23]. Hu, et al. also supported the use of the intrinsic molecular classification by distinguishing 306 genes which could not only predict the distinct molecular subtypes, but also form strong conclusions on each subtype's overall survival rate [22].
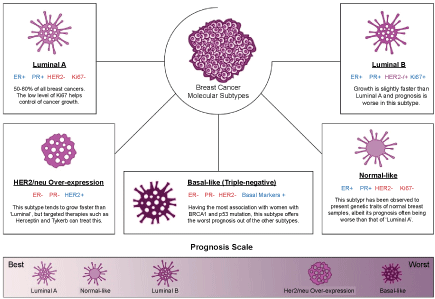 Figure 1: Molecular subtypes of breast cancer and their relative prognostic outcome. View Figure 1
Figure 1: Molecular subtypes of breast cancer and their relative prognostic outcome. View Figure 1
The 5 molecular subtypes groupings are also characterized by the range of breast cancer 1 (BRCA1) and other oncogenic mutations which may be harboured differently in each subtype [24]. Aside from that, the molecular subtypes shown in Figure 1 can also be utilized as a prognostic tool, whereby breast cancer of the luminal subtypes which account for more than 50% of total cases are lower grade and present better prognosis as compared to the triple-negative subtype. Given the absence of 3 major receptors and a high correlation with breast cancer 1 (BRCA1) mutations, the options of treatment for such patients are limited to a combinatorial radiation and chemotherapy or targeted therapies for non-HER2/neu patients [24].
There are however, differing opinions of some research groups around the world have allowed more classifications to surface, include the basal and non-basal subtypes of breast cancer or the molecular pathway classification [25,26]. Despite there are novel outlooks to molecular groupings of breast cancer, however this could negatively impact the development of treatment as there is no standardized method to predict the different classes [27]. The molecular subtypes are not only distinguishable via extensive laboratory-based microarray tests [21-23], but also other sophisticated genetic models which are available to diagnose the different types of breast cancer, such as MammaPrint [28] and Oncotype DX [29]. The availability of commercial molecular subtyping allows the prediction of a patient's cancer to a certain treatment and this prognostic power, if replicated correctly and efficiently to patient's own tumour-derived organoids will most likely increase the treatment success rate.
Having been studied extensively, various breast cancer cell lines and animal models have emerged throughout the years, each with its own distinctive traits. From the widely used MCF-7 cells to in vivo dog models, each model comes with series of advantages and limitations which are discussed in the following sub-sections. Despite the common usages of these models, it has become apparent that generic cancer models do not work optimally to predict each and every patient's conditions, let alone outcome [30].
The emergence of 2D breast cancer cell lines could be traced back to 1958 whereby the first cultivated breast carcinomas named BT-20 was developed [9,31]. The pioneering culture study allowed various studies globally to come up with their own cell lines. To date, there are over 50 different cell lines have been established in the field of breast cancer research, some of which are widely and more commonly used in the research setting [32]. It is to be considered that these cancer cell lines may represent more than 1 particular molecular subtype or to a deeper extent, epigenetic profiles [33], while others were designed to recapitulate the hallmarks of particular subtypes.
MCF-7 cells have been widely utilized for the purpose of breast cancer drug discovery as well as the pathophysiological studies of breast cancer. Various studies have demonstrated that MCF-7 cells do in fact, belong to the luminal A subtype [33,34]. Despite majority of the claims that MCF-7 belongs to just 1 subtype, there have been questions rising on the validity of the molecular grouping, as it seems that other molecular subtypes exist within cultures of MCF-7 cells, namely triple-negative [35]. The triple-negative subtype of MCF-7 seemed to develop upon culturing of MCF-7 cells with fulvestrant [35], a type of hormone receptor positive treatment of breast cancer, which raises more questions on the possible morphological and molecular changes within stable breast cancer cell lines in the face of certain treatment options. One study conducted by Jiang, et al. [36] have found that protein expression levels of patient-derived tumour samples were more reproducible not in MCF-7 cells, but in the less commonly used T47D and BT483, again underlining the idea that one cell line does not fit all.
As opposed to the generic breast cancer cell lines, the use of primary cell cultures derived from patient's own tumour have been studied to give better prognostic power [37,38], however, discrepancies could be observed within cell lines that belong to the luminal subtype as they are being passaged. This phenotypic drift often favours the competitive growth of the more aggressive basal or triple-negative subtypes, would skew the results of drug testing on such cells, thus the use of at most, first-passage cells is recommended [36,37]. The spontaneous rise of triple-negative cells in this scenario could present problems of its own as there have been very few successful treatment options for triple-negative breast cancer patients despite the availability of generic cell lines which faithfully present the molecular markers of triple-negative breast cancer cells [39], which might give false negatives in studies which focus on another breast cancer subtype other than the triple-negative.
The use of 2D cell lines, be it derived from patient's own tumour or the use of commercially established cell lines, presents a good outlook from an economic standpoint as well as in reducing excessive usage of animal models [40]. Given that the transcriptomic and proteomic profiles do match the patients' tumours behaviour, the utilization of well-characterized cell lines could allow a faster and higher throughput screening of drugs [40]. The main limitations, however, include the several key factors within cancer biology that these 2D cell cultures do not portray or adhere to, including the stromal and matrix component of cancer out of the various other microenvironment factors [41,42]. More specifically in the case of generic cell lines, some studies have pointed out that established cancer cell lines display more genetic mutations and aberrations to those found in patient-derived tumours [30,43,44]. Cancer cell lines are also prone to cross-contamination and the use of such cell lines could lead to catastrophic outcomes if not validated [45].
The use of animal models within biological research is as common as the use of cell lines. The main argument for a cancer study to utilize animal models would be that whole organisms could display the complexity of intricate diseases like cancer which gives the research a better understanding compared to the use of in vitro 2D cell cultures.
Having been utilized for various other diseases, mice models of breast cancer have been utilized copiously within the field. The clear evidence that a more robust and cost-effective diagnostic model is needed can be further emphasized by the slow emergence of functional therapies or the translational effects in patients' cases, with only lesser than 10% of drugs being screened daily making it past early phases of development [11,46]. To test the drug trastuzumab's response in alone, a meta-analysis conducted by Chen, et al. in 2016 has revealed at least 80 mice model studies using over 2000 mice in the sets of experiments [47]. Out of the thousands of preclinical mice models, many different variations exist, each bearing their own sets of benefits and drawbacks.
In syngeneic models, breast cancer cell lines are allografted within immunocompetent hosts. The main importance of syngeneic models in cancer research is the intact immunity that is present, unlike in immune-deficient models, and this pose huge implications especially in targeted therapies which are immunological-based [48]. Despite the absence of tumour resection from syngeneic such as one conducted by Bailey-Downs, et al. in BALB/cj mice [49], it has been noted that other studies have reported a significant immunogenic reaction which leads to regression of tumour growth [48]. Other important limitations are the short timespan for treatment in syngeneic models as some tumours exhibit 100% development of the primary tumour, albeit being very aggressive [49,50], and the murine allografts which are different than human-derived tumour cells and thus, species differences might occur [51].
As a combinatorial effort to attain better portrayal of the disease, breast cancer cells might also be embedded as xenografts within mice models. The cells being xenografted into the immune-deficient models can be of immortalized human breast cancer cell lines or patient-derived xenografts (PDX), and these allow the recapitulation of tumour microenvironment at a more specific level, given that it is grown within a whole organism [48,50]. In the use of immortalized cell lines however, there has been a notable lack of heterogeneity that can be observed in the case of patient-derived tumour xenografts [51]. Some of the widely studied xenografts are the HER2/neu mouse models, such as PDX in non-obese diabetic severe compromised immunodeficiency (NOD/SCID) mice in a study conducted by Liu, et al. [52] and NOD/SCID gamma (NSG) mice conducted by Iorns, et al. [53], both of which studied the metastatic type of breast cancer.
Lastly, in vivo models of breast cancer may undergo genetic modifications, producing transgenic models termed as genetically modified mice models (GEMM) which allow the induction of tumour development within the original tissue of the organism itself. Much like the syngeneic models, the biological models' immune system will remain and the GEMMs can be induced by the mouse mammary tumour virus (MMTV) [54] or using doxycycline-inducible promoters [55]. The most limiting factor in developing GEMMs for breast cancer is that it is time-consuming and costly [48,56]. Recent molecular developments however, such as RNA interference (RNAi) via short hairpin ribonucleic acid (shRNA) [57] and clustered regularly interspaced short palindromic repeats caspase 9 (CRISPR/Cas9) systems [58,59] allow a more rapid induction of such genetic changes, some of which are even applied in constructing robust organoid models discussed in later sections.
Generally, animal models require more capital to maintain and takes longer to give the desired research output as compared to 2D cell cultures, albeit being more thorough and robust. Factors such as difficulties in obtaining quantitative data and animal stress also comes into play with the use of animal models [41], with the latter also correlating to human stress to a worse prognosis in metastatic cancers. Cancer occurs at varying heterogeneity histologically and genomically between mouse models and human tumours [60], and thus the use of a more practical in vitro model is preferable and required for a higher throughput screening of drugs aside from various other applications.
Organoid cultures have been used in cancer research to model gastrointestinal [61], lung [62], or even prostate cancers [63]. This section will review the recent advances in developing breast cancer organoids alongside their uses and other limitations. Table 1 at the end of this section summarizes relevant studies and findings in breast cancer organoid research while Table 2 summarizes the advantages, limitations, and possible solutions pertaining to breast cancer organoids.
Table 1: List of current researches being done on breast cancer organoids as study models. View Table 1
Table 2: Advantages, limitations, and reported solutions of organoids as study models. View Table 2
Deriving their methods from experimental studies and modified protocols of organoid generation of other organs, researchers are continuously optimizing protocols to develop breast cancer organoid cultures. In this article, various sources of breast cancer organoids with relevant protocols are reviewed.
The first basic protocol for breast cancer organoid development involve the use of primary tumour samples have been developed by DeRose, et al. in 2013 [64]. The tumour samples were dissociated mechanically and chemically. The incubation time for digestion varies, whereby human-derived tumours require at least overnight incubation whilst mice graft-derived tumours require shorter time [64,65]. Digestions carried out with lower enzymatic concentrations at longer timepoints have also been observed to yield more viable organotypic cultures [66].
Once organotypic cultures have been initiated, it may take as fast as 24 h for the culture to reach 75% confluency, which is the amount recommended by Garbe, et al. to seed the cells optimally [65]. Organoids may not attach in the first seeding, thus adding Y27632 dihydrochloride or fasudil, which are stem cell survival promoters, may protect organoids from anoikis before seeding on new culture vessel if required [66,67].
These organotypic cultures might be subjected to lentivirus introduction in metastatic studies of breast cancer as transduction has been reported to be low in primary organoid cultures [64,68]. Once organoid cultures have matured or transduced, single cells are further separated from the culture by differential centrifugation followed by careful aspiration of the supernatant. The pellet is then resuspended in medium which can accommodate growth of mammary epithelial cells, such as M87 medium [64]. Matrigel at 4 ℃ was then added into the organoid suspension in the culture vessel and the vessel would then be inverted to let the Matrigel to solidify and not entrap the organoids at the bottom of the 3D structure. There is no measure of timepoint for organoid growth aside from reaching their desired confluency of more than 70%. There are however, factors that may affect speed of growth of organoids, including cell concentration and nutrition available within medium [64]. Incorporation of growth factors, which is absent in DeRose's study, may promote faster growth of organoid cultures [66].
Once established, the solidified organoid within Matrigel or other matrix could be visualized via immunofluorescence studies to observe viability, presence of breast cancer markers on the surface of organoids, including cytokeratin-14 (K14), cytokeratin-19 (K19), or p63 [66,69] with the nuclei counterstained with Hoechst or Rhodamine [66,70], which can indicate the degree of invasiveness of breast cancer, as well as presence of hormone receptors via metabolic imaging studies [71]. Walsh, et al. has successfully shown the growth of primary tumour samples obtained from xenografts and not just human samples in 2014 [71] and 2016 [14]. To a lesser extent, 2D cell lines could also be utilized as the core of these 3D organoid structures as opposed to patient-derived tissues [72], but the implication of this type of study could only be used for the study of the different molecular subtypes of breast cancer as they do not portray patient's own subtypes.
These established 3D organoids could also be utilized for in vitro drug tests, with applications discussed in later sections. Distinguishable from cell lines, primary tumour cells from human samples also grow significantly slower and thus proliferative studies might be carried out at longer periods. For maintenance and storage, organoids can be cryopreserved prior to adding medium and embedding it into the Matrigel matrix, or if organoids need to be maintained in 37 ℃ incubators for upcoming studies, medium needs to be replaced at least 3 times within 1 week [64,66].
With the CRISPR/Cas9 genome editing technique, Zhang, et al. have shown that molecular inactivation of 2 breast cancer related genes, namely Ptpn22 and Mll3 can promote tumorigenesis from mammary stem cells (MaSC), whereby the phenotype has been carried out in the formation of 3D organoid cultures [73]. These organoids were transduced via lentiviral vectors containing shRNA which was then implanted into GEMMs, promoting growth of tumours resembling HER2 overexpression molecular subtype. Another study which alters activation of Wnt and HER2/neu signaling within MaSC conducted by Jarde, et al. has shown to give promising results in growing 3D organoids within Matrigel matrix which resemble a less used subtype of breast cancer, Wnt1 [74,75].
Possessing various genes that may be altered, the coupling of genomic editing in MaSC has proven to be a viable option to generate tumours which can be utilized to grow 3D organoid structures. These organoids may even be grown for 30 or more days to develop the distinguishing factors of mammary gland and breast tissues [74], including the nodules as well as ductal and luminal vessels, which would give a better outlook on the mechanism of drugs being tested. Wu, et al. studied MaSC obtained from marmosets [76] and found that breast cancer organoids formed from these MaSC resemble the phenotype of human cancers, whereby they are solid and spherical for both luminal and basal subtypes, unlike in organoids derived from mice tumour grafts, which may yield hollow organoids for luminal subtype [77].
With similar steps to initiate organotypic cultures from primary breast tumour tissues in the previous sub-section explained by DeRose, et al. [64], the generation of organoids from normal breast tissues require normal breast samples of patients, which have been explained by Sokol, et al. in 2016 [78]. The derivation of organoids from this step require the administration of lentiviral vectors to transduce the cells to multiply much like in the development of intestinal organoids [79] and subsequent growth in hydrogel matrix alongside growth factor load. Although not applicable to study the progression of cancer in personalised cases per se, normal breast organoids have been utilized to study the extravasation of blood vessels as well as studies to compare biomarkers present in tumours and normal breast tissues [80].
Breast cancer organoids have been reported to possess ranging applications. The various applications from recent researches utilizing breast cancer organoids are reviewed in this sub-section.
The most desirable application of breast cancer organoids is their ability to predict the course and efficacy of treatment at a personalized scale. In the optical metabolic imaging (OMI) of primary tumour organoids which are ER+ and HER2- after comparison with the OMI profile of BT474 and HR6 organoids, results shown by the ranging fluorescence intensity indicate that treatment with trastuzumab does not change the OMI index, as well as varying response upon treatment of paclitaxel and tamoxifen, which can be clinically validated [71,81,82]. Moreover, combinatorial treatment of paclitaxel, trastuzumab, and XL14 to result in a significant reduction of OMI index for both cell line-derived organoids and primary tumour organoids from patient's own fresh sample.
Another research discussed by Sowder, et al. has shown yet another in vitro organoid culture which could be observed to express all the molecular subtypes of breast cancer heterogeneously and thus display different vulnerabilities towards current treatment based on the subtypes [83]. This in turn gives a more robust understanding on how organoids can be utilized to recapitulate not only the tumour microenvironment but also molecular subtypes of patient's tumours and thus increasing the prognostic value of this model. Within the various studies, it has been noted that the varying results, albeit being a few, may indicate the presence of cancer stem cells (CSC)-like cells which might confer to the primary tumour's drug resistance as well as renewal abilities [84].
A completed study carried out by Broutier, et al. focused on translating PDX organoids into the field of prognostics and diagnostics [85], a project which could lead to ease the pipeline within breast cancer drug discovery and to minimize the cost and time taken for these treatments to reach patients. Once established, this could be an upgraded form of the currently existing breast cancer explant biobank studied by Bruna, et al. [86], which harbours hundreds of PDX models, capable of undergoing high-throughput drug screens as well as comparative studies with the normal breast tissue samples of the same patient. This model, despite not being organoid-based, is very much like a colorectal cancer biobank constructed by van de Wetering, et al. [87], which could be the next step by incorporating organoid models in place of breast cancer PDX models alone in the biobank.
Lastly, for multiplex drug testing, researchers have come up with tumours on a chip, which is yet another innovative take on diagnostics and drug screening methodologies, whereby Torisawa, et al. [88] and Kim, et al. [89] showed protocols to create 3D cultures of breast cancer cells suspended on microchips with microfluidic channels. Cytotoxicity results of different anticancer treatments such as mitomycin C (MMC) and 5-fluorouracil (5-FU) have been reportedly comparable when grown on this chip and in MTT assay [90], although it was also observed that the growth rate of the 3D cell culture was markedly slower than 2D cell culture.
The discovery of key molecular determinants of breast cancer is important, especially in the face of genomic editing techniques, CRISPR/Cas9 or shRNA in correcting these aberrations and possibly offering us with a treatment outlet. Breast cancer organoids generated from MaSC transplanted in GEMM have provided a molecular outlook on inactivation of cancer-related genes via the CRISPR/Cas9 method which appear to be mutated in human breast cancer cases in general, namely Ptpn22 and Mll3 [73]. Compared to 2D cell lines whereby high throughput drug screens are mostly performed at, some genetic aberrations are over-represented while not being so in 3D cultures.
Within a gene expression array study, it is emphasized that signal transducers and regulatory enzyme activities are statistically over-represented in 2D cell lines and not in 3D cell cultures when compared to in vivo model [91]. The research group study also found that despite the molecular subtypes playing a major role in gene expression profiles, there still is importance of how tumour microenvironment and culture conditions affecting genomic profiles significantly [91]. Biomarker discovery in the end, still proved to be a necessary field in breast cancer research as there has been reported various shifts at a molecular level to another subtype upon 1 type of treatment regimen [35], not to mention the scarce treatment option for aggressive triple-negative basal like cancers.
We first consider the patient-derived organoids as an outlet to study human disease progressions which are otherwise difficult to study in animal models. As compared to 2D cell lines, organoids provide a more complete and thorough picture of the progression of the disease, including phenotypic similarities and biomarker profile when compared to patient's own tumours [92]. Three-dimensional cell cultures of breast cancer also show faster timepoint to gather quantitative data as compared to animal studies [14,41,64,71], and the in vitro properties of organoids makes it a more practical choice in immunofluorescence studies [14,71].
Organoids offer a biological model which can be derived from various sources of tissues, including normal tissues or tumours from adult patients, as well as embryonic stem cells (ESC) and induced pluripotent stem cells (iPSC), whereby when propagated to its optimum potential, will portray the disease alongside its microenvironment component at a high degree. It has been portrayed that fibrinogen and fibroblast components produced by metastatic breast cancer can cross react with polyethylene glycol (PEG) to form hydrogel, thus, the solid and stiff nature that organoids can grow into may also indicate the degree of metastatic breast cancer in experimental models [92,93]. Organoids grown for extensive period are stable such as shown by Jarde, et al. retained their phenotypes for up to 70 days since establishment of the mammary organoid culture, although the cell architecture seems to be lost accompanied by accumulation of chromosomal aberrations after 112 days upon establishment [74].
One of the major concerns of growing organoids as a study model in cancer research is that the loss of oncogene drive due to the growth factor-rich environment which are dubbed enough to sustain the fastened proliferation [37]. In the case of the protocol that was discussed vastly by DeRose, [64], however, the Matrigel is devoid of these growth factors and thus, such limitations should not hinder the oncogenic drive. Despite mimicking most cellular architecture and surface markers faithfully, 3D organoids have been reported to have weak vascular organization which leads to hypoxia and subsequent activation of genes which modulate drug sensitivity alongside cell survival [94]; however, several studies have indicated that this drug resistance mechanism occurs within in vivo models as well, providing organoids with natural tumour behaviour to avoid over-representation of drugs upon screens [95]. Other limitations include cost and difficulty to scale-up aside from the rigidity of the ECM component and types of scaffold used [95,96], which might indicate invasiveness or dampen incoming drug molecules, thus affecting drug tests. The future of implantation of genetically corrected organoids into humans as a therapeutic agent may also be affected by Matrigel that is widely used in the organoid scaffold structure, as it is of murine origin [97], and thus use of other types (e.g. hydrogel) might be preferable in the future use in human transplantation.
Much like live tissue or embryonic stem cell samples, patient-derived organoids such as ones deposited in live biobanks [87] come with a series of possible ethical issues. The first ethical issue is the consent requested from the patients, whether it would be a broad, tiered, or specific consent. With the novel nature of organoid research, patients might feel justified with a broad consent [98], whereby they can withdraw their participation completely, or with tiered consent whereby they can choose what their tissues can be used for from a range of research outcomes [99]. Privacy is another issue whereby the institute receiving the tissue samples should always implement strategies to keep patient information confidential. Furthermore, property rights shall be considered, as given that the tissue comes from patients, but it will subsequently undergo modifications that it might not be considered as patient's own tissue anymore, a question asks about the proprietor of the submitted tissue itself [100]. Ethics play a huge role in any case of tissue submission; thus, research institutes or bodies should come up with strategies to mediate the ethics that come into play so that the advancements in organoid studies will not be hindered.
The development of a breast cancer organoid biobank is indeed an interesting prospect, allowing patients who require diagnostic or prognostic procedures done to do so with more ease. Given the heterogeneous nature of breast cancer itself, researchers have yet to come up with a generalized protocol to generate patient's primary tumour organoids and thus more studies should be conducted. The future of organoids, if coupled with genetic modification techniques such as the CRISPR/Cas9, can offer viable therapeutic options in different instances. Study has shown that epithelial organoids generated from gastrointestinal stem cells could be functionally repaired by genetic modification to revert the effects of a single locus mutation within the cystic fibrosis transmembrane conductor receptor (CFTR) [101]. This opens possibilities of the clinical applications of genetically corrected organoid cultures to be implanted in patient tissues to correct diseases, one of which could be breast cancer.
It has become apparent that despite the glories that 2D and in vivo models have provided us, there is a dire need for a more robust translational model to study breast cancer. With current researches abiding by different protocols to develop various types of breast cancer organoids, this model offers versatile approaches to develop it, but there still needs to be a general protocol to optimize production and yield of organoid culture as a guideline. The current applications of 3D cultures of breast tumours range from multiplex drug screening to predicting the course of treatment in a personalized case of a patient, and with the dawn of genomic editing techniques and its similar applications in colorectal cancer or gastrointestinal cancer organoids, researchers might even couple these techniques to form a pioneering therapeutic option for breast cancer.