International Journal of Virology and AIDS
T-reg are the Predominant CD4+ T-Cell Subset Productively Infected with HIV-1 at Sites of Dual HIV/TB Infection
Christina S Hirsch1, Shigou Liu1, Joy Baseke2, John Lusiba Kafuluma3,4, Mary Nserko3,4, Harriet Mayanja-Kizza2-4 and Zahra Toossi1,5*
1Division of Infectious Diseases, Case Western Reserve University, USA
2Joint Clinical Research Center, Kampala, Uganda
3Makerere University, Kampala, Uganda
4National Tuberculosis and Leprosy Program, Kampala, Uganda
5Veterans Affairs Medical Center, USA
*Corresponding author: Zahra Toossi MD, Department of Medicine, Division of Infectious Diseases, Case Western Reserve University, Euclid Ave, Cleveland, OH, USA, Tel: 216-368-0441, Fax: 216-368-2034, E-mail: zxt2@case.edu
Int J Virol AIDS, IJVA-2-005, (Volume 2, Issue 1), Research Article; ISSN: 2469-567X
Received: October 30, 2014 | Accepted: February 05, 2015 | Published: February 10, 2015
Citation: Hirsch CS, Liu S, Baseke J, Kafuluma JL, Nserko M, et al. (2015) T-reg are the Predominant CD4+ T-Cell Subset Productively Infected with HIV-1 at Sites of Dual HIV/ TB Infection. Int J Virol AIDS 2:005. 10.23937/2469-567X/1510005
Copyright: © 2015 Hirsch CS A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: Regulatory T-cells (T-reg) are expanded during active tuberculosis (TB) regardless of HIV-1-infection, particularly at sites of M. tuberculosis infection. In HIV-1 disease, T-reg are targeted by HIV-1 infection. However, whether they contribute to promotion of HIV-1 infection at sites of HIV/TB is unknown.
Methods: Pleural fluid mononuclear cells (PFMC) from HIV/TB patients with pleural TB were characterized by immunostaining and FACS analysis for surface markers CD4, CD127, CCR5, CXCR4 and intracellular expression of Foxp3, HIVp24, IFN-g and Bcl-2. T-reg purified by immunomagnetic bead separation were assessed for HIV-1 strong stop (SS) DNA expression by real-time PCR.
Results: High numbers of T-reg (defined as CD4+Foxp3+ or CD4+Foxp3+CD127 –), that were HIV-1-infected were found in PFMC. T-reg displayed higher expression of the cellular activation marker, HLA-DR (p< 0.001), and HIV-1 co-receptors (CCR5 and CXCR4) (p< 0.05 for both) as compared to non-T-reg. Purified T-reg exhibited higher HIV-1 infection, as measured by HIV-1 SS DNA, when compared to whole PFMC, and as compared to PFMC T-reg from an HIV-infected subject with mesothelioma. HIV- infected CD4+Foxp3+T-reg were significantly higher in Bcl-2 expression as compared to CD4+Foxp3-cells (p< 0.001). Further, HIV-1-infected T-reg showed significantly higher Bcl-2 reactivity than un-infected T-reg (p< 0.001). A small fraction of HIV-1-infected T-reg were also IFN-g+.
Conclusion: At sites of pleural HIV/TB infection higher frequencies of PFMC T-reg with survival advantage are predisposed to productive HIV-1 infection as compared to non-T-reg.
Introduction
Expansion of CD4 T-cells that express Foxp3 is a feature of active tuberculosis (TB). High numbers of Foxp3+ CD4 T-cells with a T-regulatory functional profile in blood [1,2] and at sites of Mycobacterium tuberculosis (MTB) infection [3,4] have been documented. The basis of expansion of T-reg at sites of dual HIV-1 and TB (HIV/TB) co-infection has been studied more recently. Expression of Foxp3, a molecular signature marker of T-reg [5], among pleural fluid mononuclear cells (PFMC) at sites of HIV/TB co-infection was significantly higher than that in autologous PBMC [6]. Other has reported expression of CD39 in addition to Foxp3 as a marker of T-reg in TB [7]. Pleural sites of active HIV/TB infection are prominent in HIV-1 activity as compared to that found systemically [8], and currently activated CD4 T-cells are believed to be the predominant source of HIV-1 in situ [9]. Whether T-reg contribute to productive HIV-1 infection at sites of HIV/TB is not known.
Foxp3 expression is necessary in establishing the regulatory T-cell lineage both for thymus-derived natural (n) T-reg, and induced (i) T-reg derived from naive CD4 T-cells outside of the thymus (Reviewed in [10]). Expression of Foxp3 in iT-reg is under control by epigenetic regulatory elements [11]. In peripheral lymphoid tissues, transforming growth factor beta (TGF-β) is critical to induction of expression of Foxp3 in T-cell receptor (TCR) activated naïve CD4 T-cells [12,13]. Whereas induction of iT-reg requires TGF-β alone,expansion of TGF-β induced iT-reg is dependent on the NFkb/c-Rel [14] and/or IL-2/Stat5 signaling [15] pathways. At sites of pleural TB, TGF-β and pro-inflammatory cytokines (IL-8, IL-6, and IFN-g), but not IL-2, are abundant [6]. Further, Foxp3 mRNA expression in PFMC correlated with levels of IL-6, IL-8 and TGF-β in pleural fluid [6], suggesting that continuous TCR activation through cytokines in the cellular micro milieu at sites of pleural HIV/TB infection may be conducive to stabilization of Foxp3 expression in PFMC T-reg.
T-reg have been found to be targets of HIV-1 infection among blood mononuclear cells [16,17]. Also, naive T-cell precursors of Foxp3 positive T-reg are susceptible to HIV infection in vitro [18]. Both heightened status of cellular activation [19] and increased expression of HIV-1 co-receptors [17] may underlie predisposition of T-reg to successful viral infection. Further, Foxp3 enhances NFkb occupancy at HIV-1 LTR [20], indicating that T-reg are predisposed to productive HIV-1 infection. However, others have shown that transfection of Foxp3 gene in CD4 T cells in fact represses HIV-1 transcription [21,22]. Therefore, the transcriptional basis of HIV-1 infection in T-reg remains controversial.
Other studies have shown that CD4+Foxp3+ T-cells express IFN-g, when activated under a Th1 cytokine polarizing environment [23,24], and a Th1 cytokine milieu characterizes sites of pleural TB [6,25]. Expression of IFN-g among T-reg during HIV/TB co-infection is not known.
Based on our previous observations of both enhanced HIV activity and increased frequencies of T-reg at pleural sites of HIV/TB we hypothesized that T-reg expanded at pleural sites of dual HIV/TB infection may directly contribute to expansion of HIV-1 activity. We found that among CD4+ T-cells in PFMC, HIV-1 infection was enriched in the CD4+Foxp3+CD127-T-cell population. Higher numbers of CD4+Foxp3+CD127- T-cells co-expressed HIV-1 p24 and IFN-g, as compared to non T-reg. Expression of both HIV-1 co-receptors (CXCR4 and CCR5) and HLADR was significantly higher among PFMC T-reg than non-T- reg. HIV-1 infected CD4+Foxp3+T-cells were also Bcl-2 positive, implicating their survival advantage when compared to HIV-1 infected non T-reg. Further, HIV infected T-reg were significantly more BCl-2 positive than uninfected T-reg. Therefore, among CD4+T-cells at sites of HIV/TB co-infection, the predominant population of productively HIV-1 infected T-cellsare T-reg.
Methods
Study subjects
Patients hospitalized at Mulago Hospital at Makerere University in Kampala Uganda with symptoms of fever, cough, night sweats, dyspnea for at least 2 weeks who had moderate to large pleural effusion were identified and referred to the TB clinic of the TB Research Unit for evaluation of pleural TB. Informed consent (approved by the Institutional Review Board at CWRU and the Ugandan National AIDS Research Subcommittee) was obtained from all subjects. All patients underwent HIV-1 testing, and thoracocentesis and pleural biopsy as previously described [26]. Diagnosis of pleural TB was based on positive culture of sputum and/or pleural fluid, and/or positive histology of pleural tissue for MTB. Only HIV+ patients were included in this study (n=16). All patients received standard short-course anti-TB therapy and were followed as before. Table 1 describes the characteristics of enrolled patients.
![]()
Table 1: The characteristics of enrolled patients
View Table 1
Preparation of cells and separation of T-reg from PFMC using magnetic beads
PFMC were prepared by FicollHypaque (Pharmacia Fine Chemicals, Piscataway, NJ) density gradient centrifugation as described [26]. Viability was > 98% as assessed by trypan blue exclusion. By immmunostaining and FACS analysis PFMC were 40-60% CD4 T-cells and contained only 1-5% CD14+ macrophages.
In a few experiments, T-reg were prepared by magnetic bead separation of PFMC using the Human regulatory T-cell isolation kit II from Miltenyi Biotech (Auburn, CA) as instructed by the manufacturer. Purity of CD4+Foxp3+T-cells obtained exceeded 90%. Aliquots of whole PFMC and purified T-reg were, dissolved in Tri-reagent (Molecular Research Center, Cincinnati, OH) and stored at -70°C until use.
Analysis of cell phenotype by flow cytometry
At the onset of study 4 color flow cytometry was used, as this was the limit of the flow cytometer available on site. Antibody and isotype control antibody combinations used to characterize T-reg included: 1. CD4 PercpCy5.5, HLA-DR APC, CD127 Pac Blue and rat FoxP3 PE; 2. CD4 PercpCy5.5, IgG2a APC, IgG1 Pac Blue and rat IgG2a PE (all from eBioscience, San Diego, CA); 3. CD4 PercpCy5, CCR5 FITC, CXCR4 APC (both from Biolegend, San Diego, CA) and Foxp3 PE; 4.CD4 PercpCy5, IgG2a FITC, rat IgG2a APC and rat IgG2a PE; 5. CD4 PercpCy5, p24 FITC (Beckman Coulter, Brea, CA), Bcl-2 PE(BD Biosciences, San Jose, CA) and FoxP3 e-fluor660; 6.CD4 PercpCy5, IgG1 FITC, IgG1 PE and rat IgG2a e-fluor660 (eBioscience).
Subsequently, a MiltenyiMACSQuant Flow cytometer (allowing for the assessment of samples containing a maximum of 8 fluorochromes) became available. To assure that staining with FoxP3 antibody alone accurately identifies T-reg, subsequently duplicate samples for each patient were stained with CD127 Pac Blue (or IgG1 isotype antibody in combination with FoxP3 PE or Foxp3 alone (as above). We chose CD127 as an additional marker for T-reg as studies by Fazekas et al. and Seddiqui et al. [27,28] indicate that addition of CD127 to Foxp3 and/or CD25 allows for the identification and isolation of a pure population of T-reg and allows for a better discrimination between T-reg and activated (CD25+) T-cells. We established that a strong correlation between frequencies of T-reg characterized as CD4+Foxp3+CD127- and CD4+Foxp3+ (R=0.87) existed. Antibody combinations for 5 color experiments were 1. CD4 PercpCy5.5, CD127 Pac Blue, FoxP3 PE, p24 FITC and IFN-g Alexa Fluor 647 (Biolegend); 2. : CD4 PercpCy5.5, IgG1 Pac Blue, Rat IgG2a PE, IgG1 FITC and IgG1 Alexa Fluor 647 (Biolegend).
The protocol and buffer set from eBioscience were used for all experiments involving intracellular staining. Samples were fixed and acquired within 1h of completion of staining. Data were analyzed in bulk using FlowJo software (TreestarInc, Ashland, OR) at the completion of study.
Measurement HIV-1 DNA by real-time PCR
Cellular DNA was extracted from the lower phase of cell lysates in Tri-reagent according to the manufacturer's instructions. Real time PCR using the Taqman methodology by ABI StepONEPlusthermo cycler (Applied Biosystems, Foster City, CA)was performed for HIV-1 minus-strand strong stop (SS) DNA (which represents HIV-1 transcripts post-cell entry that have initiated reverse transcription) [29], and human beta globin DNA. In each sample DNA copies were normalized to the copy number of beta globin (0•5×1010 copies in 1 million cells).
Statistics
Normally distributed data sets were analyzed by student t-test. Wilcoxon or Kruskall Wallis tests were used for data sets that were not normally distributed. Correlation between variables was assessed using linear regression and correlation or spearman rank order correlation as appropriate. P≤0.05 was considered significant
Results
Preferential HIV-1 infection of CD4+Foxp3+IFNg+ PFMC
In combination immunostaining studies we assessed HIV-1 p24 and IFN-g reactivity of CD4+ Foxp3+/- T-cells that were negative or positive for IL7 receptor (CD127) expression. This approach fully differentiates “True T-reg” (CD4+Foxp3+ CD127-) from non-T-reg (CD4+Foxp3-CD127+) and the “transitional” population (CD4+Foxp3+CD127+) [27,28]. An algorithm of analysis ofHIV-1 p24 and IFN-g reactivity in CD4+Foxp3+CD127-and CD4+Foxp3-CD127+ is shown in Figure 1A. Cumulative data for 6 experiments is shown in Figure 1B. Higher frequencies of CD4+Foxp3+CD127- T-cells stained positive for HIV-1p24 (7.8-fold; P< 0.01) (Figure 1B left panel) when compared to CD4+FoxP3-CD127+pleural T-cells. A small % of CD4+Foxp3+CD127+ were also identified (Figure 1A).
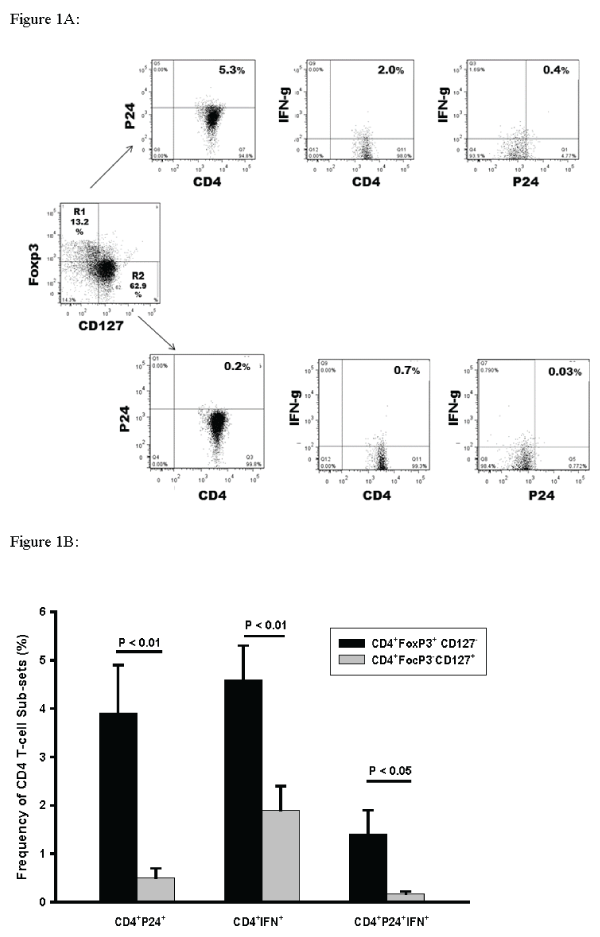
Figure 1: Frequencies of CD4+Foxp3+ CD127- and CD4+Foxp3-CD127+ PFMC co-expressing HIV-1 p24 alone or in combination with IFN-g.
True T-reg (CD4+Foxp3+ CD127-) and non-T-reg (CD4+Foxp3-CD127+) among PFMC from HIV/TB patients were characterized for their expression of HIV-1 p24
and IFN-g. A. Algorithm for assessment of frequencies of CD4+Foxp3+ CD127- and CD4+Foxp3-CD127+ PFMC co-expressing HIV p24 and IFN-g alone or in
combination. B. Among CD4+Foxp3+ CD127- PFMC frequencies of HIV-1 p24+ (p< 0.01), IFN-g+ (p< 0.01) and dual HIVp24+IFNg+ (P< 0.05) T-cells were higher
as compared to CD4+Foxp3-CD127+ T-cells (n=6 for all comparisons).
View Figure 1
We also found that IFN-g positivity among CD4+Foxp3+CD127- T-reg was 2.2-fold higher when compared to CD4+FoxP3-CD127+ non-T-reg (P< 0.01) (Figure 1B mid panel). Further, greater frequencies of CD4+Foxp3+CD127- T-reg were dually HIV-1 p24+ and IFN-g+ as compared to CD4+FoxP3- CD127+(8.2-fold; P < 0.05) (Figure1B right panel).
Productive HIV-1 infection of CD4 T-cells is dependent on cellular activation status. Therefore we also assessed the percentage of CD4+Foxp3+ CD127- and CD4+Foxp3-CD127+ PFMC T-cells that were HLA-DR+. HLA-DR positivity among CD4+Foxp3+ CD127-T-reg was 3 fold higher (paired t-test) as compared to CD4+Foxp3-CD127+ (p< 0.001) non T-reg (data not shown). Frequencies of HLA-DR+CD4 PFMCT-cells positively correlated with frequencies of Foxp3+p24+T-reg (R=0.60, p=0.01).
Expression of HIV co-receptors by CD4+Foxp3+PFMC
HIV-1 infection is dependent on expression of HIV-1 co-receptors (CCR5 and CXCR4) by CD4+ T-cells. In prior observations we had found that PFMC T-cells characterized by high CD25 expression had significantly higher expression of CCR5 as compared to CD25 low or negative CD4 T-cells (unpublished data).
In experiments outlined above (n=6) a strong correlation between frequencies of T-reg characterized as CD4+Foxp3+CD127- and CD4+Foxp3+(R=0.87) was found. Here we examined both CCR5 and CXCR4 expression by CD4+Foxp3+PFMC as compared to CD4+Foxp3- PFMC. Figure 2A shows these results: expression of both CCR5 and CXCR4 was higher on CD4+Foxp3+ as compared to CD4+Foxp3- T-cells (P< 0.01 for both comparisons, n=5).
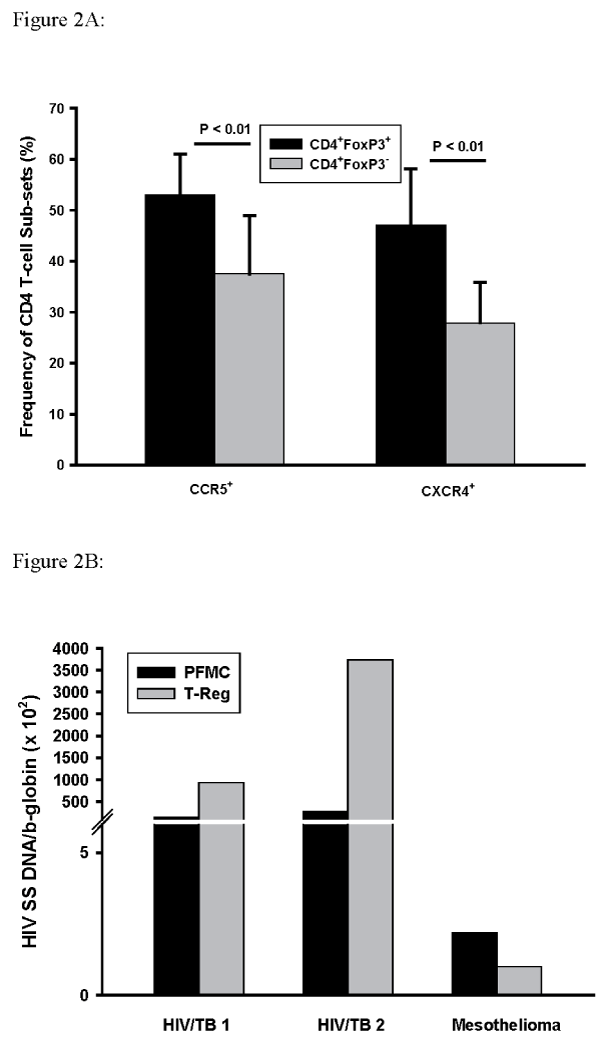
Figure 2: Expression of CCR5 and CXCR4 by CD4+Foxp3+ T-reg and of CD4+Foxp3- PFMC.
PFMC from HIV/TB patients were assessed for CD4, Foxp3+, and the
HIV-1 co-receptors CCR5 and CXCR4. Expression of CCR5 and CXCR4
was increased in CD4+Foxp3+ T-reg as compared to CD4+Foxp3- PFMC
(P< 0.01 for both, n=5) (A). HIV-1 entry was assessed by PCR following
immunomagnetic separation of PFMC to obtain T-reg (CD4+Foxp3+ CD127-
) and compared to that in whole PFMC. Results shown are for 2 HIV/TB
patients and one HIV infected subject with pleural mesothelioma (B).
View Figure 2
To confirm enrichment of HIV infection among PFMC T-reg, whole PFMC and T-reg (CD4+Foxp3+ CD127-) purified by immunomagnetic separation were compared. Cellular DNA was assessed by HIV-1 strong stop (SS) DNA (HIV-1 transcripts that have initiated reverse transcription). Results from two HIV/TB subjects were compared to PFMC and T-reg obtained from an HIV-infected subject with pleural mesothelioma. HIV-1 SS DNA was enriched among PFMC T-reg as compared to whole PFMC in HIV/TB subjects (Figure 2B). HIV-1SSDNA in PFMC from the subject with mesothelioma was low and not enriched in their T-reg.
HIV infected Foxp3+ T reg cells have survival advantage among PFMC
Previously we have observed a survival advantage (by Tunel assay) of T-reg over non-T-reg among PFMC in HIV/TB co-infected subjects (unpublished data). Here, the intracellular co-expression of Bcl-2 and HIV-1 p24 among CD4+Foxp3+and CD4+Foxp3- T-cells was assessed (n=16). A 5 fold higher Bcl-2 expression among T-reg as compared to non-T-reg was found (P< 0.001; Figure 3A). Interestingly, there was a correlation of Bcl-2 reactivity and Fox p3+ HIV-1 p24+dual reactivity (R=0.551, p=0.02), but not with HIV-1 infected non-T-reg (Fox- p24+). Interestingly, further analysis showed that expression of Bcl-2 reactivity was significantly enriched amongHIV- infected as compared to HIV uninfected Foxp3+T-reg (p< 0.001) (Figure 3B).
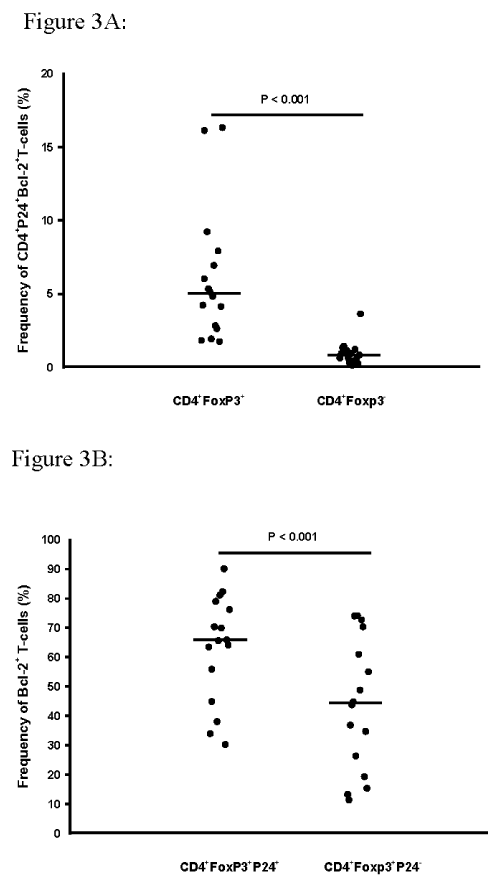
Figure 3: Survival advantage of HIV-1 infected Foxp3+ T reg cells.
PFMC from HIV/TB patients were assessed for co-expression of intra-cellular
Foxp3, HIV-1 p24 and Bcl-2 (n=16). A. Frequencies of CD4+Foxp3+ T-reg
co-expressing both HIV-1 p24 and Bcl-2 exceeded those of CD4+Foxp3-
PFMC by more than 5 fold (p< 0.001). B. Intracellular expression of Bcl-2
was enriched among Foxp3+HIV p24+, as compared to Foxp3+HIV p24- CD4
T-cell subsets were determined by simultaneous staining with antibody to
Bcl-2 (n=16).
View Figure 3
Discussion
Recent literature indicates that CD4+Foxp3+T-reg are predisposed to HIV infection [16,18] and thus may contribute to HIV-1 disease progression through viral production. This is in addition to the role of T-reg in suppression of antigen-specific CD4 and CD8 anti HIV-1 T-cell responses [30]. Here we found significantly higher numbers of T-reg defined as CD4+Foxp3+ CD127 –that were HIV-1 infected in PFMC at sites of pleural HIV/TB. T-reg were characterized by both significantly higher cellular activation (HLA-DR) and expression of HIV-1 co-receptors (CCR5 and CXCR4) as compared to non-T-reg, underscoring their susceptibility to productive HIV-1 infection. Interestingly, HIV-1 infected T-reg were Bcl-2 reactive, implicating their survival advantage not only over non-T-reg, but also over HIV-uninfected T-reg. Also, a significantly greater fraction of T-reg were both HIV-1 infected and IFN-g+, when compared to non-T-reg. Cumulatively, these data pose T-reg as a dominant HIV-infected CD4 T-cell population with survival advantage at sites of TB. Whether increased frequencies of HIV-infected T-reg directly contribute to HIV production at sites of HIV/TB through HIV release or infection of other mononuclear cells needs to be established in future studies.
During in vitro infection of T-cells by either HIV-1 [17,18] or FIV [31], T-reg support higher viral replication than non-T-reg. Both degree of HIV-1 co-receptor expression and activation status determines successful productive HIV-1 infection of mononuclear cells. Previously, we had found higher expression of both CCR5 [29] and CXCR4 (unpublished) by CD4 T-cells in PFMC from dually infected HIV/TB subjects. Here we identify CD4+Foxp3+ T-cells as the subset with significantly higher expression of CCR5 and CXCR4 as compared to CD4+Foxp3- non-T-reg in PFMC. Consistent with increased HIV-1 co-receptor expression by T-reg, we found higher levels of initiated viral transcripts (HIV-1 SS DNA) in immuno-magnetically isolated PFMC T-reg as compared to un-separated PFMC. Further, HIV-1 SS DNA was notable in T-reg from two HIV/TB patients as compared to a HIV-1 infected subject with mesothelioma.
In the current study, we found more PFMC T-reg that were dually positive for HIV-1 p24 and Bcl-2 than non-T-reg. Even among T-reg, HIV infection was associated with significantly higher expression of Bcl-2. Therefore, HIV-1 infected T-reg at sites of HIV-1/TB are presumably characterized by lesser predisposition to apoptosis as compared to other T-cells. However, others have shown that among Foxp3 targeted genes in iT-reg, expression of Bcl-2 is actually down-regulated [32], and inhibition of Bcl-2 expression appears to be based on interaction of Foxp3 with the transcriptional repressor, Eos [33]. While it is possible that at sites of HIV/TB, interaction of Foxp3 and Eos is defective in PFMC T-reg, this needs to be investigated. On the other hand, TCR engagement leads to expression of anti-apoptotic molecules Bcl-2, Bfl-1 and Bcl-xl in T-cells [34,35]. Strong Th1signaling provided at sites of HIV/TB infection [6,25], in the presence of continuous TGF-β signaling necessary in maintaining Foxp3 expression [36], likely underlies the basis of expanded Bcl-2+ T-reg among PFMC found here. The higher Bcl-2 expression of HIV-1 infected Foxp3+ T-cells and the correlation of Bcl-2 with Foxp3+p24+(and not Foxp3+p24-) reactivity underscores survival advantage of HIV-1 infected T-reg. Thus, infected T-reg at sites of HIV/TB may be the predominant CD4+T-cell source of HIV-1 due to their survival advantage. This may explain the correlation of PFMC Foxp3 mRNA to viral load in the pleural fluid of HIV/TB patients found here (data not shown, n=16, r=0.780, p< 0.002). Of note, however, there was only a modest and insignificant correlation of Foxp3 mRNA expression in PFMC with pleural fluid viral load in our previous study (n=10, r=0.5) [6].
Recently, it has been shown that CD4+Foxp3+ T-cells can produce IFN-g when activated under a Th1 cytokine polarizing environment [23,24]. In fact, it has been suggested that IFN-g production identifies pathogen epitope-specific T-reg during viral infections [37]. Here, up to 6.1% of T-reg (CD4+Foxp3+CD127- T-cells) were IFN-g+. IFN-g production by T-reg may be important to their immunoregulatory function as shown in the model of Graft-versus-Host Disease [38]. However, the true role of IFN-g producing T-reg at sites of dual HIV/TB remains to be deciphered.
In summary, higher frequencies of T-reg as compared to non-T-reg are HIV-infected and display survival advantage among PFMC at sites of pleural HIV/TB. Whether expanded T-reg at sites of dual HIV/TB infection contribute to HIV-1 viral activity also by undermining anti-HIV-1 immune responses of CD4 and CD8 effector T-cells needs to be further studied.
Acknowledgements
This study was supported by NIH grants HL 51636 and AI-1080313, and the Tuberculosis Research Unit (AI-70022). None of the authors have a commercial or other association that might pose a conflict of interest.
References
-
Scott-Browne JP, Shafiani S, Tucker-Heard G, Ishida-Tsubota K, Fontenot JD, et al. (2007) Expansion and function of Foxp3-expressing T regulatory cells during tuberculosis. J Exp Med 204: 2159-2169.
-
Singh A, Dey AB, Mohan A, Sharma PK, Mitra DK (2012) Foxp3+ regulatory T cells among tuberculosis patients: impact on prognosis and restoration of antigen specific IFN-? producing T cells. PLoS One 7: e44728.
-
Guyot-Revol V, Innes JA, Hackforth S, Hinks T, Lalvani A (2006) Regulatory T cells are expanded in blood and disease sites in patients with tuberculosis. Am J Respir Crit Care Med 173: 803-810.
-
Sharma PK, Saha PK, Singh A, Sharma SK, Ghosh B, et al. (2009) FoxP3+ regulatory T cells suppress effector T-cell function at pathologic site in miliary tuberculosis. Am J Respir Crit Care Med 179: 1061-1070.
-
Ziegler SF, Buckner JH (2006) Influence of FOXP3 on CD4+CD25+ regulatory T cells. Expert Rev Clin Immunol 2: 639-647.
-
Toossi Z, Hirsch CS, Wu M, Mayanja-Kizza H, Baseke J, et al. (2011) Distinct cytokine and regulatory T cell profile at pleural sites of dual HIV/tuberculosis infection compared to that in the systemic circulation. Clin Exp Immunol 163: 333-338.
-
Chiacchio T, Casetti R, Butera O, Vanini V, Carrara S, et al. (2009) Characterization of regulatory T cells identified as CD4(+)CD25(high)CD39(+) in patients with active tuberculosis. Clin Exp Immunol 156: 463-470.
-
Toossi Z, Johnson JL, Kanost RA, Wu M, Luzze H, Peters P, et al. (2001) Increased Replication of HIV-1 at Sites of Mycobacterium tuberculosis Infection: Potential Mechanisms of Viral Activation. J Acquir Immune Defic Syndr 28: 1-8.
-
Lawn SD, Pisell TL, Hirsch CS, Wu M, Butera ST, et al. (2001) Anatomically compartmentalized human immunodeficiency virus replication in HLA-DR+ cells and CD14+ macrophages at the site of pleural tuberculosis coinfection. J Infect Dis 184: 1127-1133.
-
Curotto de Lafaille MA1, Lafaille JJ (2009) Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity 30: 626-635.
-
Lal G, Bromberg JS (2009) Epigenetic mechanisms of regulation of Foxp3 expression. Blood 114: 3727-3735.
-
Fantini MC, Becker C, Monteleone G, Pallone F, Galle PR, et al. (2004) Cutting edge: TGF-beta induces a regulatory phenotype in CD4+CD25- T cells through Foxp3 induction and down-regulation of Smad7. J Immunol 172: 5149-5153.
-
Chen W, Jin W, Hardegen N, Lei KJ, Li L, et al. (2003) Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med 198: 1875-1886.
-
Isomura I, Palmer S, Grumont RJ, Bunting K, Hoyne G, et al. (2009) c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. J Exp Med 206: 3001-3014.
-
Corbett EL (2003) HIV and tuberculosis: surveillance revisited. Int J Tuberc Lung Dis 7: 709.
-
Moreno-Fernandez ME, Zapata W, Blackard JT, Franchini G, Chougnet CA (2009) Human regulatory T cells are targets for human immunodeficiency Virus (HIV) infection, and their susceptibility differs depending on the HIV type 1 strain. J Virol 83: 12925-12933.
-
Oswald-Richter K, Grill SM, Shariat N, Leelawong M, Sundrud MS, et al. (2004) HIV infection of naturally occurring and genetically reprogrammed human regulatory T-cells. PLoS Biol 2: E198.
-
Antons AK, Wang R, Oswald-Richter K, Tseng M, Arendt CW, et al. (2008) Naive precursors of human regulatory T cells require FoxP3 for suppression and are susceptible to HIV infection. J Immunol 180: 764-773.
-
Oswald-Richter K, Grill SM, Leelawong M, Unutmaz D (2004) HIV infection of primary human T cells is determined by tunable thresholds of T cell activation. Eur J Immunol 34: 1705-1714.
-
Holmes D, Knudsen G, Mackey-Cushman S, Su L (2007) FoxP3 enhances HIV-1 gene expression by modulating NFkappaB occupancy at the long terminal repeat in human T cells. J Biol Chem 282: 15973-15980.
-
Grant C, Oh U, Fugo K, Takenouchi N, Griffith C, et al. (2006) Foxp3 represses retroviral transcription by targeting both NF-kappaB and CREB pathways. PLoS Pathog 2: e33.
-
Selliah N, Zhang M, White S, Zoltick P, Sawaya BE, et al. (2008) FOXP3 inhibits HIV-1 infection of CD4 T-cells via inhibition of LTR transcriptional activity.Virology 381: 161-167.
-
Dominguez-Villar M, Baecher-Allan CM, Hafler DA (2011) Identification of T helper type 1-like, Foxp3+ regulatory T cells in human autoimmune disease.Nat Med 17: 673-675.
-
Wei B, Baker S, Wieckiewicz J, Wood KJ (2010) IFN-gamma triggered STAT1-PKB/AKT signalling pathway influences the function of alloantigen reactive regulatory T cells. Am J Transplant 10: 69-80.
-
Barnes PF, Fong SJ, Brennan PJ, Twomey PE, Mazumder A, et al. (1990) Local production of tumor necrosis factor and IFN-gamma in tuberculous pleuritis. J Immunol 145: 149-154.
-
Hirsch CS, Toossi Z, Johnson JL, Luzze H, Ntambi L, et al. Augmentation of Apoptosis and Interferon-gamma Production at Sites of Active Mycobacterium tuberculosis Infection in Human Tuberculosis. J Infect Dis 183: 779-788.
-
Fazekas de St Groth B, Zhu E, Asad S, Lee L (2011) Flow cytometric detection of human regulatory T cells. Methods Mol Biol 707: 263-279.
-
Seddiki N, Santner-Nanan B, Martinson J, Zaunders J, Sasson S, et al. (2006) Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med 203: 1693-1700.
-
Toossi Z, Mayanja-Kizza H, Baseke J, Peters P, Wu M, et al. (2005) Inhibition of human immunodeficiency virus-1 (HIV-1) by beta-chemokine analogues in mononuclear cells from HIV-1-infected patients with active tuberculosis. Clin Exp Immunol 142: 327-332.
-
Kinter AL, Hennessey M, Bell A, Kern S, Lin Y, et al. (2004) CD25(+)CD4(+) regulatory T cells from the peripheral blood of asymptomatic HIV-infected individuals regulate CD4(+) and CD8(+) HIV-specific T cell immune responses in vitro and are associated with favorable clinical markers of disease status. J Exp Med 200: 331-343.
-
Joshi A, Garg H, Tompkins MB, Tompkins WA (2005) Different thresholds of T cell activation regulate FIV infection of CD4+CD25+ and CD4+CD25- cells. Virology 335: 212-221.
-
Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, et al. (2007) Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445: 936-940.
-
Pan F, Yu H, Dang EV, Barbi J, Pan X, et al. (2009) Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science 325: 1142-1146.
-
Vetter ML, Johnson ME, Antons AK, Unutmaz D, D'Aquila RT (2009) Differences in APOBEC3G expression in CD4+ T helper lymphocyte subtypes modulate HIV-1 infectivity. PLoS Pathog 5: e1000292.
-
Verschelde C, Walzer T, Galia P, Biémont MC, Quemeneur L, et al. (2003) A1/Bfl-1 expression is restricted to TCR engagement in T lymphocytes. Cell Death Differ 10: 1059-1067.
-
Li C, Ebert PJ, Li QJ (2013) T cell receptor (TCR) and transforming growth factor beta (TGF-beta) signaling converge on DNA (cytosine-5)-methyltransferase to control forkhead box protein 3 (foxp3) locus methylation and inducible regulatory T cell differentiation. J Biol Chem 288: 19127-19139.
-
Zhao J, Zhao J, Fett C, Trandem K, Fleming E, et al. (2011) IFN-?- and IL-10-expressing virus epitope-specific Foxp3(+) T reg cells in the central nervous system during encephalomyelitis. J Exp Med 208: 1571-1577.
-
Koenecke C, Lee CW, Thamm K, Föhse L, Schafferus M, et al. (2012) IFN-? production by allogeneic Foxp3+ regulatory T cells is essential for preventing experimental graft-versus-host disease. J Immunol 189: 2890-2896.





