Journal of Dermatology Research and Therapy
Fluorescence in Situ Hybridization (FISH) Copy Number Abnormalities at 6p (RREB1), 6q (MYB), and 11q (CCND1) Reliably Distinguish Metastatic Versus Benign Melanocytic Lesions
Zaid Hindi*, Michael Sidiropoulos, Ayman Al Habeeb, Danny Ghazarian, Niloufar Hosseini and Kenneth J. Craddock
Department of Pathology, Toronto General Hospital, University of Toronto, Ontario, Canada
*Corresponding author:
Zaid Ben Hindi, Department of Pathology, Toronto General Hospital, University Health Network, University of Toronto, 200 Elizabeth Street, Toronto, ON M5G 2C4, Canada, Tel: 2897070045, E-mail: zaidbhindi@gmail.com
J Dermatol Res Ther, JDRT-2-017, (Volume 2, Issue 1), Original Research Article; ISSN: 2469-5750
Received: January 18, 2016 | Accepted: February 27, 2016 | Published: February 29, 2016
Citation: Hindi Z, Sidiropoulos M, Habeeb AA, Ghazarian D, Hosseini N, et al. (2016) Fluorescence in Situ Hybridization (FISH) Copy Number Abnormalities at 6p (RREB1), 6q (MYB), and 11q (CCND1) Reliably Distinguish Metastatic Versus Benign Melanocytic Lesions. J Dermatol Res Ther 2:017. 10.23937/2469-5750/1510017
Copyright: © 2016 Hindi Z, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Objectives: Malignant melanoma can be difficult to distinguish from a benign melanocytic lesion by histology. In this study, we investigated the sensitivity and specificity of FISH to distinguish between benign nevi and metastatic melanomas to lymph nodes.
Methods: Multicolour FISH was performed using a commercially available probeset (Abbott Laboratories, Abbott Park, IL), on formalin-fixed, paraffin-embedded tissue samples from 40 tumours: 20 benign melanocytic nevi, and 20 metastatic melanomas within lymph nodes, as determined by histologic assessment. Fluorescent signals for each probe were enumerated by 2 observers in 30 cells each per lesion. An algorithm using signal counts from a combination of 4 probes targeting chromosome 6p25 (containing RREB1 gene), 6 centromere (CEP6), 6q23 (containing MYB gene), and 11q13 (containing CCND1 gene) was used as suggested by the manufacturer.
Results: Of the 20 metastatic melanomas assessed, 18 were FISH positive. FISH detected significant abnormal nuclei for RREB1 in 17/20 cases (85%) and significant MYB loss in 12/20 cases (60%). Average signals per nuclei greater than 2.5 for CCND1 and MYB were present in only 7/20 (35%) and 4/20 (20%) cases respectively. All 20 benign nevi were FISH negative. Overall, the FISH test showed a sensitivity of 90% and specificity of 100% in the diagnosis of metastatic melanoma in lymph nodes.
Conclusions: These results provide further compelling evidence for the utility of multicolour FISH directed against 6p25 (RREB1), centromere 6, 6q23 (MYB), and 11q13 (CCND1) as an aid in determining malignant behavior in melanocytic lesions.
Keywords
Metastatic, Melanoma, Benign, Nevi, FISH, CCND1, MYB, RREB1
Introduction
Diagnosis of a melanocytic skin lesion as a benign nevus versus a malignant melanoma can be difficult even for an experienced pathologist. For the majority of cases, the distinction can be made based on histological features including growth patterns, mitotic activity, and nuclear atypia. For the remaining cases, however, the distinction can be very difficult due to countless histological variants and lesions with borderline features. The medical community is in need of an ancillary test that can aid in differentiating nevi from melanomas, as the speed and quality of the diagnosis can impact the patient’s prognosis and life expectancy. Mutations such as BRAF, KIT, and NRAS are not useful in diagnosis of melanomas due to their presence in benign nevi.
DNA copy number testing by comparative genomic hybridization (CGH) or fluorescence in situ hybridization (FISH) can potentially fulfill this role as an aid to the diagnosis of malignant melanoma. CGH, and more recently microarray-CGH, has shown DNA copy number abnormalities to be rare in histologically benign lesions. The exceptions to this are 11p gains (including HRAS gene) in 10-20% of Spitz nevi [1], and aneuploidies reported in giant congenital nevi [2]. The lack of gene duplication and gene deletion illustrates the intact DNA repair mechanisms in benign nevi; the dysfunction of these repair mechanisms by TP53 mutations and other mechanisms is one of the hallmarks of malignant melanomas [2]. FISH probes targeting three chromosomal regions that are commonly gained or lost in melanoma have been developed (Abbott/Vysis) and reported to have a high sensitivity and specificity for melanoma diagnosis [3-7]. The loci tested include RREB1 at 6p25 (commonly gained in melanoma), MYB at 6q23 (commonly lost), and CCND1 at 11q13 (commonly gained).
There exists limited and conflicting data regarding the sensitivity and specificity of FISH findings for certain melanocytic lesions, particularly metastatic melanocytic lesions [3,5,8,9]. The existing literature, while promising, remains inconclusive on the utility of FISH in assisting in diagnosis. This study aimed to address the gap by testing the efficacy of Abbott/Vysis FISH panel in distinguishing between benign and malignant melanocytic lesions in our laboratory, and to determine the incidence of FISH-positive status of unequivocally metastatic lesions.
Materials and Methods
Ethics approval was obtained from the UHN Research Ethics Board. Lymph node sections with metastatic melanoma deposits from 20 patients were selected to represent unequivocally malignant-behaving melanoma cells. Unequivocally benign melanocytic nevi based on histological evaluation were tested from 21 patients.
FISH was performed using an H&E circled by a dermatopathologist to guide the location of testing. After overnight heating in a dry oven at 60°C, 4 μm FFPE tissue sections are deparaffinized in 3 fresh xylene baths for 10 min each followed by dehydration in absolute ethanol for 5 min. Tissue pre-treatment was first done in 10 mM Sodium Citrate Buffer pH = 6.0 at 80°C in a water bath for 2 hours, rinsed in 2X SSC and then deionized water. Tissues were further digested in pepsin (750000 units/ml in 0.01N HCl) at 37°C in a water bath for 30 min after an initial incubation in 0.01N HCl for 10 min. Digestion was assessed under phase contrast microscopy to ensure appropriate amount of tissue digestion. Slides were further digested up to 15 min depending on the state of digestion of each case. Digestion was stopped by placing slides in deionized water for 10 min. Slides were then dehydrated in a sequential ascending ethanol series (70%, 90%, and 100%) for 2 minutes each and air-dried. Vysis Melanoma FISH probe set (Abbott Molecular, Des Plaines, Illinois) was then added to the slide (2.5 μl), coverslipped (12 mm round), sealed with rubber cement and co-denaturated using a microprocessor-controlled system (Thermobrite, Abbott Molecular, Des Plaines, Illinois) at a melting temperature of 75°C for 10 min. Hybridization was carried out at 37°C overnight. Post-hybridization washes consisted of 0.4X SSC/0.3% NP-40 at 69°C for 2 min and 0.1% NP-40/2X SSC at room temperature for 1 min. The slides were air dried in a dark chamber followed by chromatin counterstaining with DAPI/mounting medium (Vectashield medium, Vector, Burlingame, CA) and coverslipped.
As diagrammed in Figure 1, the Vysis Melanoma FISH Probe set consists of 3 probes hybridizing to chromosome 6: RREB1 located at 6p25 (Red), MYB located at 6q23 (Gold) and CEP6 (Aqua) and one probe hybridizing to chromosome 11: CCND1 at 11q13 (Green). The RREB1 (6p25) probe is labeled with SpectrumRed and covers a 638 kb region that contains the entire RREB1 gene. The MYB (6q23) probe is labeled with SpectrumGold and covers a 740 kb region that contains the entire MYB gene. The CCND1 (11q13) probe is labeled with SpectrumGreen and covers a 378 kb region that contains the entire CCND1 gene. The CEP 6 probe, labeled with SpectrumAqua, hybridizes to repetitive sequences comprising the centromere of chromosome 6 (6p11.1-q11.1).
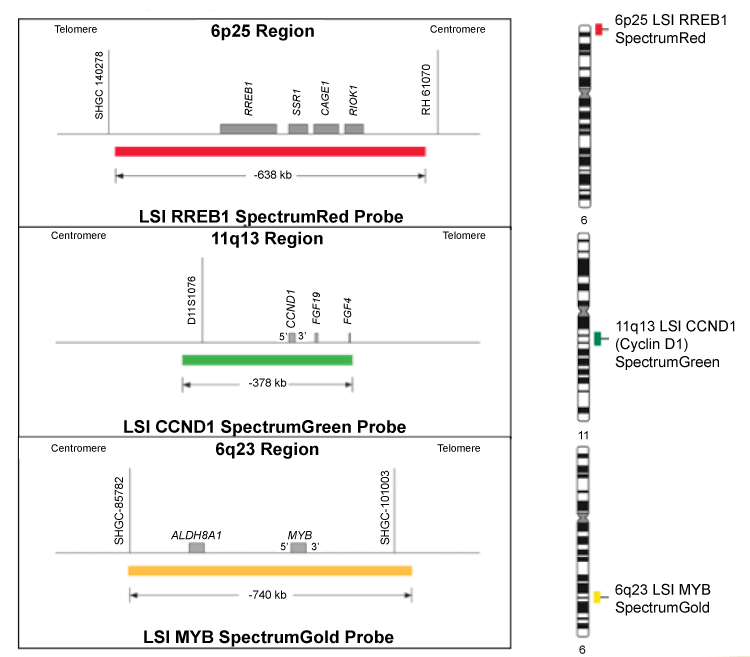
.
Figure 1: RREB1 (6p25), CCND1 (11q13), MYB (6q23) genes. The RREB1 (6p25) Probe is labeled with SpectrumRed and covers a 638 kb region that contains the entire RREB1 gene. The MYB (6q23) probe is labeled with SpectrumGold and covers a 740 kb region that contains the entire MYB gene. The CCND1 (11q13) probe is labeled with Spectrum Green and covers a 378 kb region that contains the entire CCND1 gene. The CEP 6 probe, labeled with SpectrumAqua, hybridizes to the centromere of chromosome 6 (6p11.1-q11.1).
View Figure 1
Analyses were done using a fluorescence microscope (ZeissAxio Imager, Gờttingen, Germany) equipped with a triple band-pass filter set (DAPI/Green/Orange), dual band-pass filter set (Green/Orange) and single band-pass filters (DAPI, SpectrumGreen, SpectrumGold, Aqua, SpectrumOrange & Red). Image capture was done using a digital ProgRes MF video camera (Jenoptik, Germany) and the fluorescent image acquisition software ISIS (MetaSystems, Germany).
Areas with tumor cells were circled by a pathologist on the H&E slide, and analyses were performed in corresponding areas, in a blinded fashion, by two observers using Zeiss fluorescent microscopes. The number of copies of each probe were detected and counted for thirty nuclei each, for a total of sixty tumor cells per lesion.
The manufacturer’s algorithm, included in the product insert, was applied to determine whether each lesion was FISH positive or FISH negative. The following criteria were used as per the Abbott product insert: (1) the average number of CCND1 signals per nuclei is greater than or equal to 2.5, or (2) the average number of MYB signals per nuclei is greater than or equal to 2.5 or (3) the percentage of nuclei with a loss of MYB relative to CEP6 is greater than or equal to 31% or (4) the percentage of abnormal nuclei for RREB1 (defined as any RREB1 signal count aside from two) is greater than or equal to 63%. If at least one of the four criteria were met, the specimen was designated FISH positive. It is worth noting the MYB probe has two criteria for positivity, representing either loss or gain of copy number. Although loss of MYB is the most common pattern seen in malignant melanomas, gain of copy number at this gene may also be seen in a smaller subset, and is thought to be representative of an unstable genome with many complex rearrangements resulting in copy number abnormalities manifesting as both losses and gains. If none of the above criteria were met, the specimen was designated as FISH negative. In the event of a discrepancy in FISH result between the first two observers, a third observer scored an additional 30 cells and the values given by two of the three observers, who were most in agreement, were used to give the final result.
Sensitivity of the FISH test was calculated as the percentage of malignant melanomas that were positive by FISH testing. Specificity was calculated as the percentage of benign nevi that were negative by FISH criteria.
Results
Overall, this study found the Vysis FISH panel to produce a sensitivity of 90% and specificity of 100% in the diagnosis of malignant-behaving (metastatic) melanocytic proliferations. All twenty-one benign nevi were FISH negative (Table 1) (Figure 2A and Figure 2C). Of the twenty metastatic melanomas assessed, eighteen were FISH positive (Figure 2B and Figure 2D). Most FISH positive melanomas showed abnormal RREB1 copy number (75%), and only four (25%) as a singular abnormality. Two cases were abnormal only for CCND1 copy number, and two had loss of MYB as a sole abnormality. Notably, the two metastatic melanomas that were FISH negative were actually outside of the range of counts that we found for unequivocally benign nevi for all criteria (RREB1/MYB/CCND1 counts/MYB-CEP6-relative counts), but did not reach the criteria of FISH positivity as specified in the product insert.
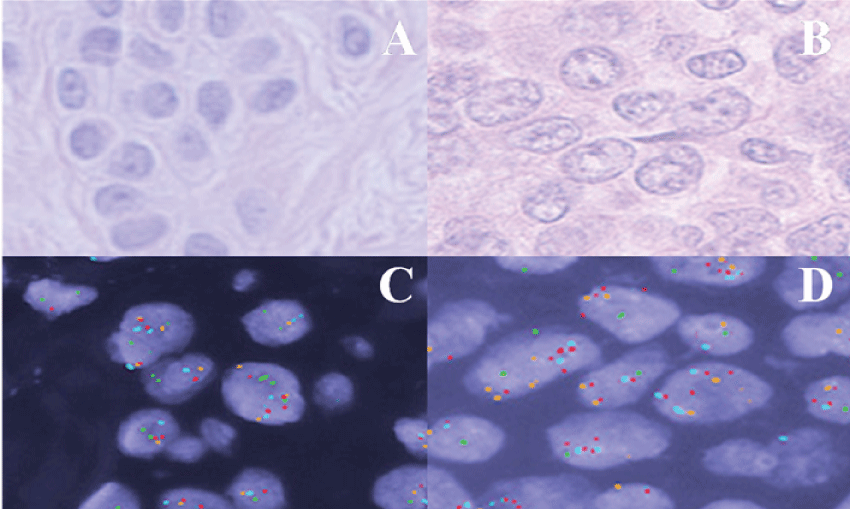
.
Figure 2: (A) H&E stained tissue sample of a benign nevus showing a nest of melanocytes; (B) H&E stained tissue section of metastatic melanoma cells within a lymph node. Melanocytes show enlarged nuclei with irregular contours and coarse chromatin; (C) Benign nevus tissue probed with melanoma FISH probe set; (D) Malignant melanoma cells probed with melanoma FISH probe set.
View Figure 2
![]()
Table 1: Summary of FISH findings for 21 benign nevi. None of the benign nevi were FISH positive.
View Table 1
Table 2 shows the detailed FISH results for each metastatic melanoma, and Table 3 summarizes the frequency of each abnormality by FISH in the metastatic melanomas. The criteria for gain of RREB1 at 6p25 was met in 14/20 cases (70%), and loss of MYB relative to CEP6 on 6q in 9/20 cases (45%). Gain of CCND1 and MYB (mean # signals ≥ 2.5) was seen in 5/20(25%) and 1/20 (5%) cases, respectively. Figure 2 shows representative images of the FISH patterns seen in benign nevi versus metastatic melanoma.
![]()
Table 2: FISH findings for the 20 metastatic melanomas. Specimens were designated FISH positive if at least one of four criteria listed in table 4 were met. Values rendering a FISH positive result are highlighted in bold.
View Table 2
![]()
Table 3: Frequency of each of the four FISH positive criteria in our 20 metastatic melanomas.
View Table 3
Neither of the scorers had a positive FISH result for any of the benign cases. Two of the metastatic melanoma cases had a discrepant FISH result from the first two scorers (M4 and M20), one of which was found by the third scorer to be FISH-negative, and the other was found to be FISH-positive. For the case that was found by the third scorer to be FISH-negative (M4), the results for the two FISH-negative scores came close to the abnormal cut-off for both RREB1 mean, and % abnormal RREB1 cells. Specifically, scorer 1 attained a RREB1 mean value of 2.4 (2.5 is the positive FISH criteria) and a % abnormal RREB1 of 50% (63% is the positive FISH criteria), while scorer 2 attained a RREB1 mean value of 3.43 and % abnormal RREB1 of 93%. Scorer 1 found 10% of the cells had an abnormal MYB-CEP6-relative count, while scorer 2 found that 73.3% of the cells had an abnormal MYB-CEP6-relative count. The third scorer found this case to be FISH negative for all criteria, with mean RREB1 of 2.45 and % abnormal RREB1 of 56%. Although this case did not quite reach the cut-offs for a FISH positive result, these RREB1 parameters are well outside the range seen in our benign nevi (Table 1). For the discrepant case that was found to be FISH positive by the third scorer, there seemed to be some heterogeneity in the tumor, with some nests of tumor cells showing increased copy numbers to the extent of meeting the FISH positive criteria, and other nests showing counts that did not meet the criteria for FISH positivity. Specifically, values for the three scorers for % abnormal RREB1 were 20%, 88%, and 80%, and average MYB copies per nucleus were 2.10, 2.47, and 2.93.
Discussion
Previous studies testing the sensitivity and specificity of FISH using this Abbott commercial probe set for the pathologic diagnosis of benign nevus versus primary malignant melanoma have largely been conducted by the same research group [4,10-13], and most of these studies focused on primary rather than metastatic melanomas. We found a sensitivity of 90% (18/20) and specificity of 100% (21/21) for this FISH probe set in the detection of malignant behavior in melanocytic lesions. In combination with three other studies that have tested FISH on metastatic melanomas [3,5,14], 58/66 metastatic melanomas have tested positive by this FISH probe set resulting in a sensitivity of 88%. This value is comparable to that which is observed in primary melanomas, ranging from 81-90% (Table 4 for a summary of previous studies and their sensitivity and specificity of FISH for pathologic diagnosis of melanoma) [3-6,8,9,14]. We can conclude from this that there is no evidence diagnostic superiority in utilizing FISH in metastatic melanomas as compared to primary melanomas. Nonetheless, FISH has proved to be a valuable tool in diagnosing these conditions.
![]()
Table 4: Summary of previous studies testing benign nevi and primary melanomas using the same FISH probe set. Sensitivity and specificity refer to pathologic diagnosis as a gold standard.
View Table 4
Numerous studies, including ours, have illustrated that 95-100% of pathologically benign lesions test negative by FISH; however, one should bear in mind that 10% of metastasizing melanomas will have a negative FISH result. It is important that the pathologist integrate findings of both histopathology and FISH in order to arrive at a circumspect diagnosis.
A major problem in this field has been in establishing the usefulness of FISH in the diagnosis of atypical or borderline melanocytic lesions such as Spitz and minimal-deviation or nevoid melanomas, and in predicting malignant behavior among difficult borderline lesions. Difficulty arises due to follow-up constraints, and the circumstance that most pathologically borderline lesions tend to be excised as a primary melanoma; therefore, an absence of metastasis does not rule out malignant potential of the original lesion.
Previous studies have found that false positive results can be obtained with this FISH probe set for Spitz nevi and other histologically borderline lesions [7]. Gerami et al. (2012) noted that the presence of polyploidy exceeding tetraploidy is a characteristic of melanoma, whereas tetraploidy may be seen in benign lesions such as Spitz nevus and congenital nevus [15]. The group utilized a multi-probe analysis analyzing 4 cohorts. The group found that a 4 probe set consisting of loci at 9p21 (CDKN2A), 6p25 (RREB1), 11q13 (CCND1), and 8q24 (MYC) to improve sensitivity and specificity. When using this 4-probe set compared to that of the one used in our investigation, the group found the sensitivity and specificity to increase from 75% and 96% to 94% and 98%, respectively. The utilization of loci 9p21 and 8q24, as demonstrated by this group, appears to improve statistical classification of the melanoma and nevi. Ultimately, however, the diagnosis of malignant melanoma still requires an overall assessment of histologic criteria in addition to FISH results, and we cannot rely completely on genetic testing to guide our determination of benign versus malignant behavior.
Conclusion
These results provide compelling evidence for the utility of multi-colour FISH directed against 6p25 (RREB1), centromere 6, 6q23 (MYB), and 11q13 (CCND1), as an aid in determining malignant behavior in melanocytic lesions. A positive result provides strong support for a malignant diagnosis, and although a negative result does not rule out malignant behavior, it can provide the pathologist some additional level of certainty in making a benign diagnosis when used in parallel with histological findings. FISH can aid in timely diagnosis, improving prognosis by allowing the medical unit to diagnose, stage, and treat the disease at an earlier state.
It is important to be mindful of the different types of melanocytic lesions encountered in daily practice. Future studies may consider the study of various melanocytic lesions in utility of FISH to determine how the diagnostic criteria can be configured to optimize the above mentioned classification functions. Our future goal as a collective is to produce a standardized method utilizing FISH with appropriate diagnostic criteria as to reduce the false negative rate and increase the false positive rate. Developing an efficacious technique will aid the medical unit in reducing morbidity and mortality, while increasing patient life expectancy. The diagnostic criteria can be improved upon to maximize the effect on sensitivity and specificity. As exhibited by Gerami et al. (2012), investigation into variable probe sets and their utility in the continuum of melanocytic lesions could benefit the FISH technique as a diagnostic tool [7,16]. FISH utility in melanoma is a relatively new tool requiring improved diagnostic criteria.
Disclosure
None of the coauthors in this study have any conflict of interest.
References
-
Bastian BC, LeBoit PE, Pinkel D (2000) Mutations and copy number increase of HRAS in Spitz nevi with distinctive histopathological features. Am J Pathol 157: 967-972.
-
Calonje JE, Brenn T, Lazar AJ, McKee PH (2011) McKee’s Pathology of the Skin: Expert Consult - Online and Print 2 Vol Set, (4th edn), A Saunders Title.
-
Gerami P, Jewell SS, Morrison LE, Blondin B, Schulz J, et al. (2009) Fluorescence in situ hybridization (FISH) as an ancillary diagnostic tool in the diagnosis of melanoma. Am J Surg Pathol 33: 1146-1156.
-
Gerami P, Mafee M, Lurtsbarapa T, Guitart J, Haghighat Z, et al. (2010) Sensitivity of fluorescence in situ hybridization for melanoma diagnosis using RREB1, MYB, Cep6, and 11q13 probes in melanoma subtypes. Arch Dermatol 146: 273-278.
-
Morey AL, Murali R, McCarthy SW, Mann GJ, Scolyer RA (2009) Diagnosis of cutaneous melanocytic tumours by four-colour fluorescence in situ hybridisation. Pathology 41: 383-387.
-
Newman MD, Lertsburapa T, Mirzabeigi M, Mafee M, Guitart J, et al. (2009) Fluorescence in situ hybridization as a tool for microstaging in malignant melanoma. Mod Pathol 22: 989-995.
-
Gerami P, Li G, Pouryazdanparast P, Blondin B, Beilfuss B, et al. (2012) A highly specific and discriminatory FISH assay for distinguishing between benign and malignant melanocytic neoplasms. Am J Surg Pathol 36: 808-817.
-
Gaiser T, Kutzner H, Palmedo G, Siegelin MD, Wiesner T, et al. (2010) Classifying ambiguous melanocytic lesions with FISH and correlation with clinical long-term follow up. Mod Pathol 23: 413-419.
-
Vergier B, Prochazkova-Carlotti M, de la Fouchardière A, Cerroni L, Massi D, et al. (2011) Fluorescence in situ hybridization, a diagnostic aid in ambiguous melanocytic tumors: European study of 113 cases. Mod Pathol 24: 613-623.
-
Dalton SR, Gerami P, Kolaitis NA, Charzan S, Werling R, et al. (2010) Use of fluorescence in situ hybridization (FISH) to distinguish intranodal nevus from metastatic melanoma. Am J Surg Pathol 34: 231-237.
-
Gerami P, Barnhill RL, Beilfuss BA, LeBoit P, Schneider P, et al. (2010) Superficial melanocytic neoplasms with pagetoid melanocytosis: a study of interobserver concordance and correlation with FISH. Am J Surg Pathol 34: 816-821.
-
Gerami P, Zembowicz A (2011) Update on fluorescence in situ hybridization in melanoma: state of the art. Arch Pathol Lab Med 135: 830-837.
-
Newman MD, Mirzabeigi M, Gerami P (2009) Chromosomal copy number changes supporting the classification of lentiginous junctional melanoma of the elderly as a subtype of melanoma. Mod Pathol 22: 1258-1262.
-
Fang Y, Dusza S, Jhanwar S, Busam KJ (2012) Fluorescence in situ hybridization (FISH) analysis of melanocytic nevi and melanomas: sensitivity, specificity, and lack of association with sentinel node status. Int J Surg Pathol 20: 434-440.
-
North JP, Garrido MC, Kolaitis NA, LeBoit PE, McCalmont TH, et al. (2014) Fluorescence in situ hybridization as an ancillary tool in the diagnosis of ambiguous melanocytic neoplasms: a review of 804 cases. Am J Surg Pathol 38: 824-831.
-
Senetta R, Paglierani M, Massi D (2012) Fluorescence in-situ hybridization analysis for melanoma diagnosis. Histopathology 60: 706-714.





