International Journal of Pathology and Clinical Research
Proficiency Assessment Mechanism for Line-Probe Assay Testing for Diagnosis of Multidrug-Resistant Tuberculosis in India
Raizada N1*, Sachdeva KS2, Malhotra B3, Thakur R1, Kishore Reddy VC1, Chauhan DS4, Dave P5, Mohammed F6, Ram Das4, Sharma P4, Sharma SK7, Wares F8, Sahu S9, O'Brien R1#, Mundade Y10, Dewan PK11 and Paramasivan CN1
1Foundation for Innovative New Diagnostics (FIND), Switzerland
2Central TB Division, Directorate General of Health Services, Ministry of Health and Family Welfare, India
3Sawai Man Singh Medical College, India
4National JALMA, Institute for Leprosy and other Mycobacterial Diseases, India
5State TB Cell, Directorate of Health Services, India
6State TB Training and Demonstration Centre, India
7Department of Medicine, All India Institute of Medical Science, India
8Global TB Programme (GTB), World Health Organization, Switzerland
9Stop TB Partnership, Switzerland
10UNITAID, Switzerland
11Office of the World Health Organization (WHO) Representative to India, India
#Currently freelance consultant, Foundation for Innovative New Diagnostics (FIND), India
*Corresponding author:
Raizada N, Foundation for Innovative New Diagnostics (FIND), India, E-mail: drneerajraizada@gmail.com
Int J Pathol Clin Res, IJPCR-1-004, (Volume 1, Issue 1), Research Article; ISSN: 2469-5807
Received: May 25, 2015 | Accepted: June 05, 2015 | Published: June 08, 2015
Citation: Raizada N, Sachdeva KS, Malhotra B, Thakur R, Kishore Reddy VC, et al. (2015) Proficiency Assessment Mechanism for Line-Probe Assay Testing for Diagnosis of Multidrug-Resistant Tuberculosis in India. Int J Pathol Clin Res 1:004. 10.23937/2469-5807/1510004
Copyright: © 2015 Raizada N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
GenoType MTBDRplus assay (LPA) has been endorsed by the World Health Organization for rapid screening of patients at risk of MDR-TB for an early diagnosis. A key constraint in the uptake of this technology in 2009 was the lack of LPA proficiency assessment system. To enable LPA scale-up, proficiency testing procedures were developed and implemented in India.
Methods: After initial LPA training, each project site collected remnant sputum specimens from 50 smear-positive patients for anonymous LPA testing. Results were assessed for negative control validity, successful amplification and internal and external reproducibility. Any site failing to achieve the pre-defined proficiency benchmarks was reviewed for their practices and subsequently was reassessed, after undertaking the necessary corrective action.
Results: Six laboratories underwent LPA proficiency assessment and this mechanism revealed several procedural weaknesses, including errors in deoxyribonucleic acid (DNA) extraction practices, reagent contamination, and inter-observer variability in interpretation. After corrective action, 5 laboratories subsequently achieved required proficiency benchmarks and started service delivery for patient care.
Conclusions: LPA proficiency assessment facilitated successful implementation of LPA in India. Based on these results, the described proficiency testing mechanism was accepted by National Laboratory Committee of India's Revised National TB Control Programme, and is being implemented at all LPA laboratories under country's laboratory scale-up plan.
Background
Globally in 2012, it is estimated that 450 000 people developed multidrug-resistant TB (MDR-TB; strains of Mycobacterium tuberculosis resistant to at least rifampicin (R) and isoniazid (H)) and there were an estimated 170 000deaths from MDR-TB [1]. However, the progress towards targets for diagnosis and treatment MDR-TB has been slow. In most countries with a high burden of MDR-TB, less than 25% of the people estimated to have MDR-TB were detected in 2012. This global threat of MDR-TB to TB control, underscores the importance of prompt and rapid identification of such resistant Mycobacterium tuberculosis (M.tb) strains. With an estimated 2.2 million new TB cases a year, India has the largest number of TB cases globally, and also has one of the largest number of estimated MDR-TB cases in the world (64,000 amongst only the notified PTB cases in 2012) [1]. Up to the time of this study, the Government of India's Revised National TB Control Programme (RNTCP) had relied on conventional solid Lowenstein Jensen (LJ) culture and drug susceptibility testing (C&DST) for the diagnosis of drug resistant TB cases.
Newly developed molecular based diagnostic methods have major advantages over conventional phenotypic methods in terms of both accuracy and turnaround time. The GenoType MTBDRplus assay is a commercially available line probe assay (LPA) from Hain Lifescience, Nehren, Germany, and is designed to simultaneously detect the most important gene mutations conferring R (rpoB genes) and H (inhA, katG) resistance in M. tb isolates [2]. Testing can be performed on culture isolates or acid fast bacilli (AFB) positive sputum specimens and can provide results within 8 hours, making this a promising tool to accelerate the diagnosis of MDR-TB cases. In 2007, the World Health Organization (WHO) endorsed the use of GenoType MTBDRplus molecular LPA for the rapid screening of patients at risk of MDR-TB in low and middle income countries [3,4].
However, an important challenge in 2008-2009 to the rapid uptake of this molecular diagnostic test was at that time the lack of available guidance on standard quality assurance mechanisms, including the criteria for the minimum proficiency benchmarks to be achieved by laboratories newly implementing the LPA prior to provision of clinical service delivery. As quality assurance procedures are crucial to ensure that laboratories are proficient in their conducting of the respective test and thus providing reliable results to providers, this was a major limitation. The high sensitivity of nucleic acid amplification assays such as polymerase chain reaction (PCR) is also a draw-back, since even the smallest amount of DNA can be amplified. Target amplification methodologies such as LPA therefore require strict adherence to a number of procedures to minimize the risk of contamination leading to false-positive results [4]. As part of a wider pilot study of LPA in India, proficiency assessment procedures for LPA testing were developed and piloted, with the objective of assessing the capability of laboratories to produce accurate and interpretable results on patient specimens and also detecting errors in laboratory results and their causes. We share here our experiences of this pilot which were later adopted and are currently used as benchmarks for nationwide LPA scape-up in India.
Materials and methods
Settings
The current study reported here was part of a larger demonstration project undertaken by the Foundation for Innovative New Diagnostics (FIND) in India to assess the feasibility of introducing LPA testing in all the six existing RNTCP public sector reference laboratories at that point of time. The reported study was undertaken in 2008-09, and in order to facilitate the feasibility assessment at the 6 functional reference laboratories providing service delivery under RNTCP, a proficiency assessment mechanism for LPA testing was developed and piloted in the 6 laboratories. These laboratories were already providing MDR-TB diagnostic services for the RNTCP using LJ C&DST. However, these reference laboratories had no prior experience in the implementation of LPA. In these laboratories, specimen processing for culture was already being performed in biological safety cabinets (BSCs) in at least Biosafety Level (BSL) 2 facilities. Under the study, different rooms with restricted access for DNA extraction, preparation of re-agents for PCR, PCR amplification and hybridization, and interpretation of results (post-amplification) were provided. Further, each room was provided with separate set of equipment and unidirectional work-flow was established to reduce the likelihood of amplicon contamination. Each laboratory was equipped with -20C deep freezers for storage of DNA extracts and PCR products.
LPA proficiency assessment mechanism
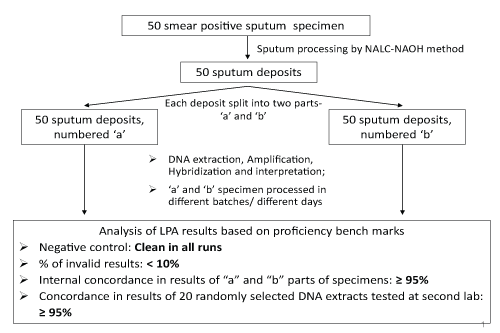
Remnant specimens of 50 fresh smear positive sputum specimens were collected from the RNTCP designated microscopy centers in the vicinity of the respective reference laboratory. Each of the specimens were assigned a study number and processed by the conventional N-acetyl-L-cysteine-sodium hydroxide (Nalc-NaOH) method (with final NaOH concentration of 1%) as recommended by the US Centers for Disease Control and Prevention [5,6]. The aliquot of these processed sputum deposits was split into two parts, 'a' and 'b' for the LPA test. From here on, the 'a' and 'b' parts were processed in separate batches of 5-10 specimens per batch for LPA by DNA extraction, PCR amplification, and hybridization, leading to 100 DNA extracts, PCR products and LPA test result strips from 50 'a' and 'b' parts of sputum deposit (Figure 1). For the 100 LPA tests conducted on 50 'a' and 'b' parts of sputum deposit, each batch of LPA testing included one negative control to rule out cross-contamination and one positive control that served as an indicator to the overall quality of all bands. The presence of any bands in the negative control, apart from conjugate control and amplification control, was interpreted as probable contamination of the entire run. In such cases, procedures required an investigation of contamination source, and a repeat of the entire run starting from the DNA extraction procedure.
The piloted proficiency assessment mechanism comprised of procedures to assess four key aspects of LPA testing, namely: (a) contamination;(b) proficiency of producing valid results; (c) internal reproducibility of results; and (d) external reproducibility of results. Each laboratory where LPA testing was being introduced participated in a five day hands-on training organized by FIND on standard operating procedures (SOPs) as recommended by the manufacturer [2].
The 50 pairs of results were locally compared for internal lab reproducibility. Once LPA testing results for both "a" and "b" aliquots of all 50 patients specimens were available, high-resolution scanned images with interpretation were e-mailed to an external umpire. The external umpire reviewed the results for LPA band interpretation errors; if any interpretation errors were found, capacity building exercises for the lab staff were conducted and continued until lab staffs were able to accurately interpret LPA results as per manufacturer recommendations.
The external umpire subsequently selected 20 of the 100 available LPA results for blinded LPA re-testing at a second LPA lab to establish external reproducibility. DNA extracts stored at -20°C, of these 20 specimens were sent to an identified external reference lab for repeat LPA testing. The external umpire evaluated all results from the labs for the following indicators, with pre-defined proficiency benchmarks (Figure 1). Invalid LPA results were defined as specimen bands staining lighter than amplification control bands, or missing bands of conjugate control or M. tb (TUB) control.
After initiation of service delivery for internal quality check of LPA results, controls were incorporated into every LPA run. A negative control (a specimen with no DNA added) and a positive control (using a suspended culture isolate with known susceptibility results, subject to the same DNA extraction procedures, amplification, and hybridization) were included in each LPA run. For external quality assurance, the procedures described earlier were repeated at 6 months and one year after initially achieving proficiency benchmarks, and repeated annually thereafter.
Laboratory procedures
To extract DNA, 0.5ml of processed sputum deposit was centrifuged at 10000g for 15 minutes, re-suspended in 100μl of molecular grade water, sealed and heated for 20 minutes at 95°C in a water bath followed by ultrasonication for 15 min at room temperature. This suspension was centrifuged at 13000g for 5 minutes, and the supernatant (DNA Extract) transferred by pipette to a fresh tube without disturbing the pellet. The Genotype MTBDRplus assay version.1 was used in the study [2].
Amplification was performed by combining 35 μl of primer nucleotide mix (PNM) with 5μl of 10× PCR buffer (containing 15mM MgCl2), 2μl MgCl2 (25 mM MgCl2), 3μl molecular grade H2O, 0.2μl (1 unit) Hot-Star Taq polymerase (QIAGEN, Hilden, Germany), and 5μl of the DNA for a total final volume of 50.2 μl. The amplification profile for direct patient material as described by the manufacturer was used for all sputum specimens. First, the template DNA was denatured for 15 minutes at 95°C, followed by 10 cycles consisting of 30s at 95°C and 2 minutes at 58°C, with an additional 30 cycles consisting of 25s at 95°C, 40s at 53°C and 40s at 70°C. The final cycle consisted of an 8 minute run at 70°C.
Hybridization was performed manually using a Twincubator, preheated to 45°C. Twenty micro liters of denaturation solution were mixed thoroughly in a plastic 12-well tray with 20μl of amplified sample (PCR product) and incubated at room temperature for 5 minutes. One milliliter of hybridization buffer was added to each trough and mixed. One pre-labelled test strip was placed into each well, and incubated the test strips and solutions for 30 minutes at 45°C. All solutions were completely aspirated following incubation. One milliliter of stringent wash solution was then added to each strip and incubated for 15 minutes at 45°C. Once all solutions were completely aspirated, and 1mL of rinse solution was added to each strip for 1 minute. The rinse solution was then completely removed; 1mL of diluted conjugate was added to each strip, and incubated for 30 minutes. After incubation, all solutions were removed, and the test strips were rinsed twice using a rinse solution for 1 minute, followed by distilled water for 1 minute. All solutions were completely aspirated between rinses. One mL of diluted substrate was then added to each strip and incubated the test strips protected from light for up to 5-7 minutes. All solutions were removed, and the reaction was stopped by rinsing twice with distilled water. The test strips were allowed to dry, and then taped to the LPA worksheet for interpretation.
Each Genotype MTBDRplus line probe strip consists of 27 reaction zones (bands), including six controls (conjugate, amplification, M. tb complex locus controls for rpoB, katG, and inhA), eight rpoB wild-type (WT) and four mutant (MUT) probes, one katG wild-type and two mutant probes, and two inhA wild type and four mutant probes. Results were interpreted according to the manufacturer's instructions [2]. The remnant processed specimen deposits, DNA extracts and PCR products of all samples were stored at -20°C. Each laboratory was equipped with -20°C deep freezers for storage of DNA extracts and PCR products separately in different freezers; The DNA extracts and PCR products were systematically labeled and stored in deep freezers and were available for proficiency related retesting at a later date.
Ethics statement
The pilot study was conducted in India, based on a Memorandum of Understanding between FIND and the Government of India (GoI) for the introduction of rapid new TB diagnostics into the RNTCP. Patients were managed as per the organizational policies and guidelines of RNTCP, based on the results of smear microscopy and solid C&DST.
At the time of the pilot study under the auspices of RNTCP, the performance of LPA had not yet been validated in the country setting and was not endorsed by RNTCP for the care of patients. Hence the LPA results were not made available to the clinicians nor considered for decision making on patient care as per the national guidelines. As per the project protocol, blinded testing (without any identifiers on the patient's specimens being available) was conducted on remnant specimens for quality assurance, which otherwise would have been discarded. Thus the results had no influence on patient care and management. As per the study protocol, no informed consent was therefore required in the study. The study protocol was reviewed, including the relevant ethical aspects, and approved by the National Laboratory Committee of the RNTCP.
Results
The results of the pilot are described in Table 1. Overall, 5 of the 6 laboratories that began proficiency assessment successfully achieved proficiency benchmarks; 1 laboratory (Lab D) dropped out after one round due to high contamination and improper storage of reagents. Only 2 of the 6 laboratories achieved proficiency benchmarks on the first attempt. Three laboratories were successful is achieving the proficiency benchmarks on the 2nd or 3rd attempt after retraining. Five out of 6 laboratories that achieved proficiency benchmarks subsequently commenced LPA testing for clinical care.
![]()
Table 1: Results of Line probe assay proficiency testing pilot
View Table 1
The follow-up and evaluation of procedures conducted in the 3 laboratories that failed to achieve proficiency benchmarks, detected various errors. As LPA was introduced for the first time in these laboratories, inaccurate interpretation of banding pattern was a common identified issue. This was addressed mostly by one on one interaction for capacity building of the concerned laboratory staffs. Most instances of contamination were due to limited prior experience with molecular testing and poor laboratory technique pre-disposing to molecular cross-contamination which was promptly identified by this proficiency assessment mechanism. Instances of contamination were attributed to improper handling while pipetting specimen and due to improper implementation of uni-directional workflow leading to carry over contamination by DNA being introduced into the clean laboratory areas. Inadequate concentration while processing the specimen by NALC-NaOH method and pipetting errors lead to suboptimal DNA yield, causing instances of high proportion of invalid results. Under the study, no correlation was observed between invalid test results and sputum smear grading. Other common errors noted in the laboratories were pipetting errors and deviation from SOPs during DNA extraction and Hybridization procedures. These errors were corrected by retraining the laboratory staff on the lab SOPs with special focus on objectively identified shortcoming in the previous proficiency assessment.
Discussion
While molecular methods have considerable advantages for scaling up programmatic management of drug resistant TB by offering rapid results and potential for high throughput [7], the care of patients with tuberculosis (TB) starts with a quality assured diagnosis [7]. In the context of the global drive to scale up diagnostic and treatment services for MDR-TB, rapid molecular tests such as LPA will play an increasingly important role in early detection and start of treatment. With the expanding availability of these low cost rapid tests for the detection of rifampicin resistance both in public and private sector the quality of test results being provided routinely needs to be ensured. Easy to implement standard quality assurance procedures are crucial to ensure that laboratories are proficient in laboratory procedures and are providing reliable results to providers, so that patients with MDR-TB are accurately detected and those without MDR TB are not subject to unnecessary expensive and potentially toxic treatment.
Recently, the Global Laboratory Initiative and WHO have issued guidance on LPA QA, using panel testing with artificially spiked sputa with heat killed bacilli for rapid assessment of performance of a laboratory [8]. However at the time of the study in 2008-2009, globally-recommended quality assurance protocols for LPA for MDR-TB were not yet available, and hence the standard proficiency assessment approach reported here was developed to facilitate scale-up of quality assured LPA laboratory services in India.
This LPA proficiency assessment mechanism was implemented in all the 6 existing public health laboratories. The failure of the initial proficiency assessment round in 4 out of 6 laboratories, and the subsequent identification of serious procedural errors, indicated the value of the proficiency assessment processes. The process provided the laboratory staff adequate opportunity to practice LPA testing before proceeding to have their results used for patient care, and provided the programme and the laboratories simple standard quantitative indicators to assess the quality and reliability of testing in a given laboratory. These findings gave laboratories and supervisors valuable experience in addressing the encountered errors like cross-contamination and internal discordance and a good understanding of various future technical monitoring needs. The mechanism provided the national programme high levels of confidence before proceeding for patient care based on LPA results, which facilitated the subsequent deployment of the new technology at a nationwide scale. The turnaround time for proficiency testing was quite short (2 to 8 weeks), and did not unduly delay the deployment of crucial drug-resistant TB diagnostic services. Though cost analysis of the piloted mechanism wasn't undertaken, the cost essentially includes both direct and indirect the costs of undertaking 100 LPA tests. These costs may vary at different settings depending on variance in lab infrastructure, staff salary, etc. However, the same may be escalated if the identified quality related issues are necessitating repetition of the procedure.
Based on the results of the pilot, the described LPA proficiency assessment mechanism was accepted by the RNTCP's National Laboratory Committee, [9] and is being implemented in the laboratory scale-up plan of India. This mechanism is currently being used in India under RNTCP for LPA proficiency assessment.
These experiences may be considered by other countries or laboratories in conjunction with other available guidance on quality assurance (such as that issued by the Global Laboratory Initiative [GLI]), for development of standard quality assurance procedures for LPA or other semi-automated nucleic acid amplification diagnostic tests for TB.
While the GLI panel testing using spiked sputa, is a simple and easy to implement, the proficiency mechanism described here offers an opportunity for the laboratory personnel ample opportunity to practice and imbibe necessary skills to ensure accurate results, including experiences in improving laboratory performance and trouble shooting. The mechanism described here might be considered by different laboratories as a stepping stone ahead on routine GLI panel testing.
Limitations
The piloted proficiency assessment mechanism doesn't take into account inter-technician variation in bench work skills. This mechanism assesses the proficiency of the laboratory as a unit. This aspect may be of a major concern where there are many technicians being routinely assigned different laboratory task. There have been some concerns raised about ability to produce valid results on DNA extracts after repeated freezing and thawing after storage at -20°C [10,11]. In the reported study, more than 600 DNA extracts were routinely stored at -20°C as on a number of occasions it was not feasible to undertake both DNA extraction and amplification on the same working day. A significant number of these frozen DNA extract were shipped to a different laboratory and showed very similar results. Hence overall in the current study the freezing and thawing of DNA extracts did not seem to impact on the ability of the laboratories to provide interpretable and comparable results.
While some studies have reported a correlation between LPA invalid results and smear grading below 1+ [12], other studies have documented no significant correlation between smear grading and LPA results [13]. Under the current study this aspect was not a specific area of focus. However, it was observed that most invalid results were related to errors in the laboratory handling of specimens and corrective action in this regard had a positive impact in reducing the invalidity rates.
With large scale up of LPA testing happening across several laboratories/countries, and large volumes of tests being routinely undertaken, both, the frequency and the number of samples included in the assessment may need to be revisited.
Conclusion
With the global impetus on scaling up diagnostic and treatment services for MDR-TB, rapid molecular tests such as LPA play an increasingly important role in early detection and start of treatment. While the expanding availability of these low cost rapid tests is rapidly being achieved in public and private sector the quality of test results being provided routinely needs to be ensured. The mechanism offers an easy to implement approach, which incorporates in its design objective assessment which can be easily be adopted even by small LPA laboratories currently not covered under any formal QA mechanism as well as large programmes leading to larger public health impact of quality assured testing, by providing larger population access to accurate result and enabling providers in making informed treatment decisions. The mechanism described here might be considered by the different laboratories as a stepping stone ahead on routine GLI panel testing.
Acknowledgement
The study reported here was part of the larger LPA demonstration project undertaken by FIND as per their memorandum of understanding with GoI. The funds for the study were covered under the grants received by FIND from Bill and Melinda Gates Foundation for the purpose of conducting demonstration studies on newer diagnostics.
We wish to acknowledge the contributions of the Dr VM Katoch, Secretary, Department of Health Research Ministry of Health & Family Welfare, GoI & Director-General Indian Council of Medical Research, the laboratory staff of all the project sites, covered under the project and the WHO-RNTCP consultants for their efforts in the introduction of rapid TB diagnostics.
NR, RT, VCKR, ROB and CNP are employed by FIND, a non-profit organization that collaborates with industry partners, including Hain Lifescience, for the development and evaluation of new diagnostic tests.
FW and PD are staff members of WHO. The authors alone are responsible for the views expressed in this publication and they do not necessarily represent the decisions or policies of WHO.
References
-
Pretorius C, Glaziou P, Dodd PJ, White R, Houben R (2014) Using the TIME model in Spectrum to estimate tuberculosis-HIV incidence and mortality. AIDS 28: S477-487.
-
Genotype MTBDRplus™, version 1.0 Product insert. Nehren, Germany: Hain Lifescience, GmbH.
-
STAG2007reportfinal5Septwithcolorcover.doc - stag_report_2007.pdf
-
WHO Policy Statement on Line Probe Assays FINAL.doc policy_statement.pdf
-
Abate G, Aseffa A, Selassie A, Goshu S, Fekade B, et al. (2004) Direct colorimetric assay for rapid detection of rifampin-resistant Mycobacterium tuberculosis. J Clin Microbiol 42: 871-873.
-
Revised National Tuberculosis Control Programme (2012) Guidelines on Programmatic Management of Drug Resistant TB (PMDT) in India.
-
World Health Organization - Information note -TB Diagnostics and Laboratory Services.
-
Training Package LPA (2012) Stop TB Partnership Global Laboratory Initiative.
-
Minutes of National lab committees of Revised National tuberculosis programme, India.
-
DNA storage and quality 2011.
-
Shao W, Khin S, Kopp WC (2012) Characterization of Effect of Repeated Freeze and Thaw Cycles on Stability of Genomic DNA Using Pulsed Field Gel Electrophoresis Biopreservation and Biobanking 10: 4-11.
-
Raizada N, Sachdeva KS, Chauhan DS, Malhotra B, Reddy K, et al. (2014) A multi-site validation in India of the line probe assay for the rapid diagnosis of multi-drug resistant tuberculosis directly from sputum specimens. PLoS One 9: e88626.
-
Yadav RN, Singh BK, Sharma SK, Sharma R, Soneja M, et al. (2013) Correction: Comparative Evaluation of GenoType MTBDRplus Line Probe Assay with Solid Culture Method in Early Diagnosis of Multidrug Resistant Tuberculosis (MDR-TB) at a Tertiary Care Centre in India. PLoS ONE 8: 10.1371.