C3orf21 (chromosome 3 open reading frame 21), also known as xyloside xylosyltransferase 1 (XXYLT1), is located on chromosome 3q29. C3orf21 belongs to the glycosyltransferase 8 family, furthermore, it is a retaining glycosyltransferase. In the extracellular domain of target proteins, XXYLT1 catalyzes addition of the second sylose, and then elongates the O-linked xylose-glucose disaccharide attached to EGF-like repeats. XXYLT1 negatively regulates Notch signaling, and aberrations in the Notch signaling pathway were associated with a few types of cancers. So, C3orf21 must be associated with cancer development. Our previous research has found that the C3orf21 polymorphisms were associated with lung cancer risk, and the ablation of C3orf21 gene (XXYLT1) promoted lung cancer MSTO-211H cell proliferation, inhibited apoptosis and accelerated cell migration. Understanding the role of C3orf21 in the cancer development may provide insights into the potential of C3orf21 as a cancer biomarker. Here, we reviewed several representative studies for understanding the C3orf21 and Notch signaling in cancer.
Biomarker, Cancer, Chromosome 3 open reading frame 21, Notch pathway, Xyloside xylosyltransferase 1
The number of cancer survivors continues to increase because of the advancements in both early detection and treatment, especially in the target therapy. Thus, further understanding of the molecular basis of cancer may lead to the identification of new molecular targets. Notch signaling pathway is one of the several other signaling pathways that have been shown to play an essential role in the development of all metazoans, and defects in the Notch pathway lead to a variety of cancers [1-4]. Notch signaling activation is modulated by the differential O-linked glycosylation of its extracellular domain (NECD) [5]. Xylose is transferred to the O-glucosylated mammalian Notch epidermal growth factor (EGF) repeats [6] and an xyloside xylosyltransferase 1 (XXYLT1) enzyme to further elongate the Xyl-Glc-O-Ser disaccharide on Notch EGF repeats [7]. XXYLT1 has conformed in the negative regulation of Notch signaling [8]. C3orf21 (chromosome 3 open reading frame 21) gene is also known as chromosome 3 open reading frame 21 and is encoded by UDP-xylose: α-xylosideα1, 3-xylosyltransferase (XXYLT1). Previous research studies also reported that C3orf21 rs2131877 polymorphism (C/C + C/T > T/T) was nominally associated with a reduced risk of lung adenocarcinoma (OR = 0.619, 95% CI = 0.390-0.976) [9]. Its ablation was significantly associated with lung adenocarcinomas with a higher degree of malignance [10]. C3orf21 may act as a susceptible marker for cancer therapy. So, our review mainly focused on understanding the role of C3orf21 in cancer development.
XXYLT1 is a type II membrane protein located in the endoplasmic reticulum (ER) with its catalytic domain protruding into the lumen [11]. XXYLT1 belongs to glycosyltransferase family 8 (GT8) [12], and is a retaining GT, i.e. the α-linked xylose in the donor UDP-xylose retains its stereochemistry after being transferred to the acceptor xylose [7]. XXYLT1 was first identified using glucoside xylosyltransferase 1 (GXYLT1) in a position-specific iterated-Blast and possessed an N-terminal signal sequence or membrane anchor and two conserved DXD motifs (Figure 1) [11]. Following research studies found that XXYLT1 have a GT-A fold with glycosyltransferase signature DXD motif (residues 225-227) [13]. Yu H, et al. found that the binary structure of XXYLT1 with acceptor Xyl-Glc-EGF modifies the newly synthesized Notch receptor with multiple tandem EGF repeats. When the crystal structure of the binary complex XXYLT1: Xyl-Glc-EGF was oriented like apo-XXYLT1 (as in Figure 2a), the acceptor substrate Xyl-Glc-EGF indeed was bound to the side of the enzyme (Figure 2b). Superimposition of the human Notch1 EGF11-13 crystal structure on the XXYLT1-bound hFA9 EGF was oriented to the EGF11-13 N-terminus away from and the C-terminus toward the membrane [14]. The full-length XXYLT1 formed an SDS-resistant dimer via the AXXXA dimerization motif in the transmembrane helix [7].
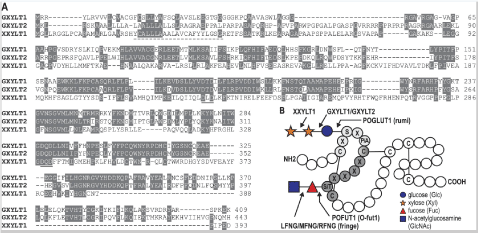 Figure 1: Sequence alignment of Notch xylosyltransferases and overview of enzymes involved in Notch glycosylation. (A) Amino acid sequence alignment of XXYLT1 with GXYLT1 and GXYLT2; (B) Overview of enzymes involved in O-fucosylation and O-glucosylation of specific consensus sequences on EGF repeats [7]. View Figure 1
Figure 1: Sequence alignment of Notch xylosyltransferases and overview of enzymes involved in Notch glycosylation. (A) Amino acid sequence alignment of XXYLT1 with GXYLT1 and GXYLT2; (B) Overview of enzymes involved in O-fucosylation and O-glucosylation of specific consensus sequences on EGF repeats [7]. View Figure 1
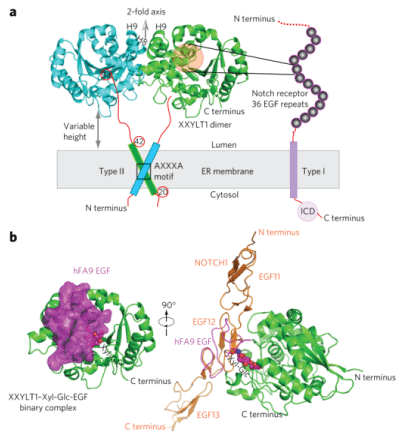 Figure 2: The mouse XXYLT1 is a dimer and has a GT-A fold with its site facing sideways to facilitate lateral modification of Notch. (a) The overall structure of type II membrane protein XXYlT1, with its truncated n-terminal transmembrane domain shown as a cylinder. The noncrystallographic two-fold axis is oriented upward and perpendicular to the sketched ER membrane in gray. The type I membrane protein notch receptor is sketched with its 36 EGF repeats as spheres. ICD, intracellular domain; H7, helix 7; H8, helix 8; H9, helix 9; Xyl-Glc, xylose-glucose disaccharide; (b) Left, overall structure of XXYlT1 in complex with hFA9 Xyl-Glc-eGF. right, superposition of the crystal structure of human noTCH1 eGF11–13 (PDb 2vJ3) with the acceptor EGF, showing that the enzyme interacts laterally with the protein substrates. XXYlT1 is in green and is in the same orientation as the green apo-XXYlT1 structure in a. Xyl-Glc-eGF is in magenta with a semitransparent surface view, and the covalently linked disaccharide is in spheres. The human noTCH1 eGF11-13 is shown in orange [14]. View Figure 2
Figure 2: The mouse XXYLT1 is a dimer and has a GT-A fold with its site facing sideways to facilitate lateral modification of Notch. (a) The overall structure of type II membrane protein XXYlT1, with its truncated n-terminal transmembrane domain shown as a cylinder. The noncrystallographic two-fold axis is oriented upward and perpendicular to the sketched ER membrane in gray. The type I membrane protein notch receptor is sketched with its 36 EGF repeats as spheres. ICD, intracellular domain; H7, helix 7; H8, helix 8; H9, helix 9; Xyl-Glc, xylose-glucose disaccharide; (b) Left, overall structure of XXYlT1 in complex with hFA9 Xyl-Glc-eGF. right, superposition of the crystal structure of human noTCH1 eGF11–13 (PDb 2vJ3) with the acceptor EGF, showing that the enzyme interacts laterally with the protein substrates. XXYlT1 is in green and is in the same orientation as the green apo-XXYlT1 structure in a. Xyl-Glc-eGF is in magenta with a semitransparent surface view, and the covalently linked disaccharide is in spheres. The human noTCH1 eGF11-13 is shown in orange [14]. View Figure 2
The XXYLT1 played an important role in the trisaccharide synthesis, which catalyzes the transfer of xylose to Xylα1-3Glc, and form D-Xylpα1-3-D-Xylpαl- 3-D-Glc, X2G. XXYLT1 elongates the O-linked xylose-glucose disaccharide attached to the EGF-like repeats in the extracellular domain of target proteins by catalyzing the addition of the second xylose [7,15,16]. XXYLT1 participated in the biosynthesis of Glc-O-type sugar chains and encodes xylosyltransferase activity [17]. XXYLT1 was active only when linked with an acceptor containing Xylα1, 3Glcβ1, while no activity was observed when linked with xylose linked to pNP or with Xyl-O-EGF16 [18]. Furthermore, XXYLT1 activity required divalent cation participation, and showed its highest activity with 20 mM Mn2+ at pH 7.2. It can also utilize Mg2+, suggesting that this might be the physiological cofactor [15]. After XXYLT1 activities, it transfers the second xylose to O-glucosylated EGF repeats of Notch [7]. XXYLT1 was involved in the organogenesis. The rearrangement of 3q29 microdeletion/microduplication syndrome region including XXYLT1 was associated to Oculo auriculo vertebral specturm [19]. XXYLT1 microdeletion also was associated with Chiari malformation type II [20]. In addition, the XXYLT1 rs9825174 polymorphism was associated with associated with osteoporosis and obesity [21], and C3orf21 rs2720932 was associated with systemic lupus erythematosus (SLE) in the Korean population [22].
The Notch signaling pathway is a short-range communication transducer that is involved in regulating many cellular processes (proliferation, stem cell and stem cell niche maintenance, cell fate specification, differentiation, and cell death) during development and renewal of adult tissues. The human Notch family includes four cell-bound notch receptors, namely Notch 1-4, and five Notch ligands, Jagged 1 (JAG1), JAG2, Delta-like 1 (DLL1), DLL3 and DLL4 [23,24]. There are 36 tandem EGF-like repeats in the Notch extracellular domain [25], they were modified by O-linked fucose, glucose, or N-acetylglucosamine (GlcNAc) [26]. Notch activation is modulated by the differential O-linked glycosylation of its extracellular domain (NECD) [20], whereby O-linked glucose is added to a subset of EGF repeats in the NECD domain by Protein O-glucosyltransferase 1 (POGLUT1) [5,27,28]. This can be subsequently extended to xylose α1-3 glucose trisaccharide by the sequential action of Glucoside α1-3 xylosyl transferase (GXYLT1/2) [17] and xyloside α1-3 xylosyl transferase (XXYLT1) [29]. GXYLT1 and GXYLT2 (glucoside-xylosyltransferase 1/2) transfer the first 1,3-linked xylose to O-glucosylated mammalian Notch EGF repeats [29], while XXYLT1 can transfer the second xylose to o-glucosylated EGF repeats of Notch [7].
Notch signaling is a juxtacrine cell-cell communication pathway with broad roles in animal development and adult tissue homeostasis [19,30]. Both gain- and loss-of-function mutations in Notch pathway components cause human disease [31-33]. Now, the Notch signaling pathway has been an attractive target in cancer research. The study found that the oncogenic effects of Notch signaling are dependent on the type of cancer: In most cases, Notch aberrations were associated with a few types of cancers [2-4]. However, in some cases such as squamous carcinomas derived from the epidermis, lung, and head and neck, and acute myeloid leukemia, inactivation of Notch signaling is oncogenic [1]. The XXYLT1 negatively regulates Notch, and reduced activity of XXYLT1 would lead to enhanced Notch signaling [8]. These results suggested that XXYLT1 gene is associated with cancer and has been conformed in larger research studies: XXYLT1 gene amplification, mutation, deletion and multiple alterations were found in several cancer types. In cancers including breast, ovarian, esophagus, head & neck, cervical, uterine, especially in lung squamous cell carcinoma, the XXYLT1 has higher amplification incidence. Notably, the activity-reducing mutations occurred in cancers with much lower incidence of XXYLT1 amplification (Figure 3), in the same time, XXYLT1 gene have 22 cancer-associated missense mutations: In human lung adenocarcinoma, the XXYLT1 mutation was A (Alanine)→E (Glutamic) (Table 1) [14]. Chromosomal imbalance at 3q29 was found in various types of cancer, including Non-small cell lung cancer (NSCLC), prostate cancer and head and neck squamous cell carcinoma [34,35]. Our previous study has demonstrated that C3orf21 polymorphism was associated with reduced risk of never smoker adenocarcinoma [9]. In Korean population, genome-wide scan found that polymorphisms at 3q29 were significantly associated with lung cancer risk, particularly with an increased risk of NSCLC. Also, C3orf21 rs2131877 was considered as the most significant biomarker of lung cancer susceptibility [36]. Our recent work also indicated that the values of NSE (Neuron Specific Enolase) tumor marker were significantly higher in patients with rs2131877 T/C + C/C genotype. In vitro study of C3orf21 mRNA expression was associated with lung cancer risk and its ablation promoted lung cancer cell proliferation, inhibited apoptosis and accelerated cell migration [10]. These studies demonstrated that C3orf21 may be a potential anticancer treatment target.
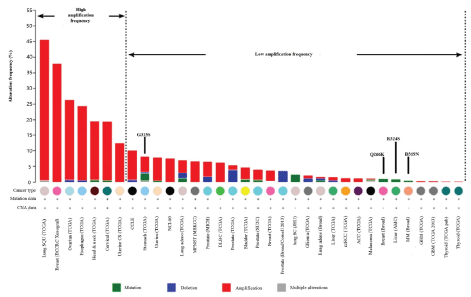 Figure 3: The abnormal expression of human XXYLT1 in difference cancers. Gene alteration data were obtained from cancer genomics site: cBioPortal [14]. View Figure 3
Figure 3: The abnormal expression of human XXYLT1 in difference cancers. Gene alteration data were obtained from cancer genomics site: cBioPortal [14]. View Figure 3
Table 1: Compilation of XXYLT1 mutations identified in some cancers. View Table 1
From the aforementioned findings, we understood that C3orf21 is one of the members of the three genes that belong to the glycosyltransferase 8 family. These were formerly named as GLT8D3, GLT8D4, and C3ORF21, but were now named as GXYLT1, GXYLT2, and XXYLT1 according to their function. C3orf21 encodes xylosyltransferases that extends O-glucose on EGF-like repeats of Notch to form the trisaccharide, Xyl-Xyl-Glc-Ser [7,29]. But XXYLT1 negatively mediated Notch signaling because of its reduced activity, which would lead to enhanced Notch signaling [14]. This in turn hinders the normal cell differentiation, resulting in epithelial cell polarity disorder and also drives normal cell to tumor cell when mature NOTCH signaling was excessively activated [37]. This suggested that C3orf21 played a significant role in tumorigenesis. In our research, we found that ablation of C3orf21 gene (XXYLT1) promoted lung cancer, MSTO-211H cell proliferation, inhibited apoptosis and accelerated cell migration. At the same time, C3orf21 overexpression showed no significant alterations in cell proliferation or apoptosis [10]. Similar phenomenon has been observed in other studies. Notch 1 protein suppressed tumor proliferation under normoxic conditions in lung cancer; however, it exhibited the opposing role of tumor promotion under hypoxic conditions [38]. Furthermore, the role of Notch 1 played different roles in different stages of tumor development. For instance, it promoted tumor development in early-stage of cervical cancer and suppressed tumor development in late-stage cervical cancer [39-41]. Whether the carcinogenesis of C3orf21 overexpression was dependent on specific tissue context, microenvironment, and crosstalk with other signaling pathways needs further elucidation. Presently, some studies regarding the role of C3orf21 in cancer development have shed greater insights in the research of C3orf21 as a biomarker of cancer [9,10,14,36]. To date, there are a few studies about C3orf21 gene, while future studies should focus on the role of C3orf21 in carcinogenesis may provide novel and in-depth understanding of the C3orf21 physiology.
This study was supported by grants from Zhejiang Provincial Science Department Public Technology Application Research Fund (#2016C33056) and Zhejiang Provincial Medical and Health Scientific and Technical Fund (#2017171306).
None.