Journal of Genetics and Genome Research
Correlation of Killer Immunoglobulin like Receptor Genes with the Rate of Cytomegalovirus Infection in Renal Transplantation Cases
Tejendra Singh Chauhan1, Swayam Prakash2, Raj Kumar Sharma1 and Suraksha Agrawal2*
1Department of Nephrology, Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow, India
2Department of Medical Genetics, Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow, India
*Corresponding author:
Dr. SurakshaAgrawal, Department of Medical Genetics, Sanjay Gandhi Post Graduate Institute of Medical Sciences, Raebareli Road, Lucknow (UP) 226014, India, Tel: 091-522 -2668004-8; Fax: 091-522 -26680973; Email: suraksha@sgpgi.ac.in
J Genet Genome Res, JGGR-1-003, (Volume 1, Issue 1), Research Article; ISSN: 2378-3648
Received: September 10, 2014 | Accepted: October 01, 2014 | Published: October 04, 2014
Citation: Chauhan TS, Prakash S, Sharma RK, Agrawal S (2014)Correlation of Killer Immunoglobulin like Receptor Genes with the Rate of Cytomegalovirus Infection in Renal Transplantation Cases. J Genet Genome Res 1:003. 10.23937/2378-3648/1410003
Copyright: © 2014 Chauhan TS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Aim: Immune mechanisms of Cytomegalovirus (CMV) infection suggest a possible relationship between CMV with development of acute graft rejection. Current immune suppression impairs antiviral specific T-cell immunity in solid organ transplantation. Inhibitory/activating NK receptor bindings activated by self HLA antigens confront allogeneic cells that lack a ligand for specific receptor. KIR ligand incompatibility caused due to presence/absence of KIR receptor in recipient and corresponding HLA ligand by the allograft which is recognized by KIR, may have potential impact on chance of CMV infection and graft survival in renal transplantation recipient. We hypothesized that predominance of activating KIR genes may downplay the rate of CMV infection among kidney transplant recipients.
Methods: We have evaluated matches/mismatches between KIR genes and known HLA ligands among CMV disease (n=27) and primary CMV infected (n=259) conditions among North Indian renal transplant cases. Sequence specific primed polymerase chain reaction method was used for KIR genotyping.
Results: Survival analysis revealed increased CMV risk for individuals carrying inhibitory KIR genes 2DL1 (OR=3.45, p-value=0.013) and 3DL1(OR=3.23, p-value=0.032), while protective association was revealed for activating KIR2DS1 (OR=0.32, p-value=0.005). Compatible KIR2DL2-HLA-C1 combination showed protective association (OR=0.23, p=0.031) with its ligand HLA-Bw4. KIR-HLA ligand match-mismatch also revealed protective association (OR=0.95, p-value=0.014) in the absence of KIR3DS1-HLA-Bw4 combination.
Conclusion: Graft outcome after renal transplantation revealed prolonged survival in the presence of certain KIR/HLA class I ligand combinations among CMV diseased and primary CMV infected cases.
Keywords
Human Leukocyte Antigen-B,C (HLA-B,C); Killer Immunoglobulin-like Receptors (KIR); Cytomegalovirus
Introduction
Cytomegalovirus (CMV) infection is the most common viral complication following solid organ as well as hematopoietic stem cell transplantation [1,2]. While specific cytotoxic T-cell immunity represents a key factor to clear CMV [3,4], very little is known about the potential antiviral role of Natural Killer (NK) cells after transplantation. Solid organ transplantation is of special interest because current immuno suppressive protocols target the activation of T cells and therefore impair specific antiviral immunity [5]. In contrast, NK cell function appears to be unaffected by such therapeutic regimens [6,7] and could therefore potentially play an important role in the anti-CMV immune response.
The clusters of KIR and HLA genes have been identified on two different chromosomes i.e., 19 and 6 respectively, leading to the possibility that an individual can lack the corresponding HLA ligand for a given KIR receptor as each can segregate independently. The balance of activating and inhibitory signals is crucial in regulating NK cell effector function, and the absence or down-regulation of MHC class I expression (missing KIR ligand) can induce susceptibility of a potential target cell to NK cytotoxicity [8,9].
The pivotal roles of NK cells and KIR receptors to eliminate the CMV virus have been highlighted in studies after human allogenic stem cell transplantation [10,11]. In addition to KIR receptors, the activating C-type lectin receptors NKG2C and NKG2D have also been shown in the defense mechanism against CMV in normal blood donors [12], and may play a critical role in viral clearance during acute CMV infection after solid organ transplantation. This suggests that NK cell phenotype and function should be considered as a significant factor when analyzing the probability of CMV infection after solid organ transplantation.
In the present study, we have compared the KIR and HLA genotypes to the risk of CMV infection in a cohort of 286 kidney transplant recipients. We have hypothesized that the absence of the HLA ligand for inhibitory KIR (i.e., missing KIR ligand) would be the case for HLA-C2, HLA-C1 and HLA-Bw6 homozygote recipients and the presence of certain activating KIRs, could protect patients from CMV infection.
Materials and Methods
We have investigated 286 cases (Males=245 (86%), Female=41 (14%)) and their respective donors (Male=60 (21%), Female=226 (79%)) in the present study. All patients and controls were residing in the state of Uttar Pradesh and adjoining states, since last three generations. Patients who have undergone renal transplantation were included in this study. All patients were maintained on triple immunosuppressive therapy that incorporated a calcinurin inhibitor (cyclosporine or tacrolimus), anti-metabolite (MMF or azathiorpine) and prednisolone. Some patients were on m-TOR inhibiter (everolimus) in place of calcinurin inhibitor. For each patient, information was collected for baseline factors like age, gender, creatinine, urinary protein level, blood urea nitrogen, blood pressure, complete lipid profile, sodium, potassium, calcium, inorganic phosphate, alkaline phosphate and tested for significance with the values obtained. DNA was extracted from blood of ESRD patients and controls collected in EDTA coated collection vials using Quiagen kits (Brand GMbH and Co KG, Cat # 51104). Quantitative CMV PCR was performed using DNA isolated from peripheral blood leukocytes to confirm the CMV status among cases. This work was approved by the ethics committees of SGPGIMS, Lucknow and Department of Biotechnology, Government of India, New Delhi. Informed consent of individuals was obtained as per the Declaration of Helsinki.
KIR and HLA genotypingAll DNA samples of transplant patients and controls were typed for KIR genes and HLA alleles basing on one of our previous study [13].
Number of matches and mismatches for KIRs with known ligands (KIR2DL1/HLA-C2, KIR2DL2/HLA-C1, KIR2DL3/HLA-C1, KIR3DL1/HLA-Bw4) and assumed HLA ligands (KIR2DS1/HLA-C2, KIR2DS2/HLA-C1, KIR2DS3/HLA-C1) have been calculated. A condition is considered to be mismatched when the recipient displays a certain KIR receptor but the donor graft does not have the corresponding HLA ligand. Similarly the case where a defined KIR receptor is expressed by the recipient and the corresponding HLA ligand is displayed by the allograft is considered to be matched. A missing condition has been assigned when the allograft has a certain HLA allele but the recipient is lacking the corresponding receptor, or when recipient and allograft both are lacking the receptor as well as the corresponding HLA ligand [14].
Statistical analysisGene frequency of KIR was determined by direct counting. Frequency differences between the CMV infected and non-CMV infected cases for individual inhibitory and activating KIR genes and the haplogroups were tested for significance at 95% confidence limits using two-tailed Fisher's exact test with Bonferroni correction. To test whether a certain KIR gene profile is associated with CMV infection in a well characterized renal allograft recipient context, binary logistic regression was applied. p-value = 0.05 was considered significant. Magnitude of effect was estimated by odds ratios and their 95% confidence intervals. Graft survival was calculated according to the principle of Kaplan and Meier using SPSS (Statistical Package for the Social Sciences software-version 16.0) and is indicated as percent ± standard error. Statistical significance was estimated using the log-rank test.
Results
Demographic and biochemical criteria
Samples were collected from patients with complete clinical details and were on a follow-up since last twelve years in the tertiary care unit of Department of Nephrology, SGPGIMS, Lucknow. Various biochemical parameters associated with renal function have been studied between CMV and non-CMV infected cases (Supplementary Table 1).
KIR gene frequencyUpon analyzing the KIR gene frequencies for their statistical associations with CMV infected cases when compared to non-CMV infected cases, we did not find any significant association of KIR genes with CMV infected renal transplant cases (Table 1). On combining HLA-B homozygous and heterozygous groups protective association with KIR3DL1 (p=0.0201, OR=0.60, 95%CI=0.40-0.92) for CMV infected cases (Table 2) has been noted.
![]()
Table 1: KIR gene frequency distribution among cytomegalovirus and noncytomegalovirus
infected renal transplantation cases .
View Table 1
![]()
Table 2: KIR receptor-HLA ligand association among cytomegalovirus and noncytomegalovirus
infected renal transplantation cases.
View Table 2
KIR/HLA ligand incompatibilities and CMV infection impact on graft outcome
Binary logistic regression was applied among CMV and non-CMV groups on the basis of KIR receptor-HLA ligand match-mismatch. Protective associations were found among cases where a CMV or a non-CMV affected individual's allograft have particular HLA allele but its corresponding receptor was absent in the recipient. Such protective associations were observed for KIR2DL2-HLAC1 (p=0.031, OR=0.23, 95%CI=0.11-0.32); KIR2DS1-HLAC2 (p=0.048, OR=0.12, 95%CI=0.09-0.31) and KIR3DS1-Bw4 (p=0.002, OR=0.46, 95%CI=0.26-0.66). Similarly protective association (p=0.014, OR=0.95, 95%CI=0.43-1.13) for CMV affected cases was also found when the patient was having KIR3DS1 but the donor graft lacked the corresponding HLA-Bw4 ligand (Table 3).
![]()
Table 3: KIR-HLA ligand matches/mismatches among CMV and non-CMV cases using binary logistic regression.
View Table 3
Survival analysis
The overall mean cumulative graft survival among CMV infected patients was 71 (95%CI=68-79) months; while the mean cumulative graft survival of non-CMV patients was 89 (95%CI=81-94) months. Level of significance was obtained for KIR2DL1-HLAC2 (?2=3.45, p-value=0.013), KIR2DS1-HLAC2(X2=0.32, p-value=0.005) and KIR3DL1-HLABw4 (?2=3.23, p-value=0.032) when the association of KIR-HLA ligand matches/mismatches on renal allograft rejection among CMV infected cases were estimated (Table 4). Subsequent analysis showed more number of CMV infected cases were on Tacrolimus (p=0.0002, OR=5.32, 95%CI=3.61-8.91) based drug regimen. While CMV cases maintained on cyclosporine revealed protective associations (p=0.0001, OR=0.76, 95%CI=0.16-0.89) (Supplementary Table 1). Prolonged cumulative graft survival has been noted for cases with KIR-Bx haplotype (Figure 1). CMV cases maintained on ATG induction showed graft survival upto 67 months.
![]()
Table 4: Effect of KIR-HLA ligand matches/mismatches on graft survival among CMV infected renal transplant cases.
View Table 4
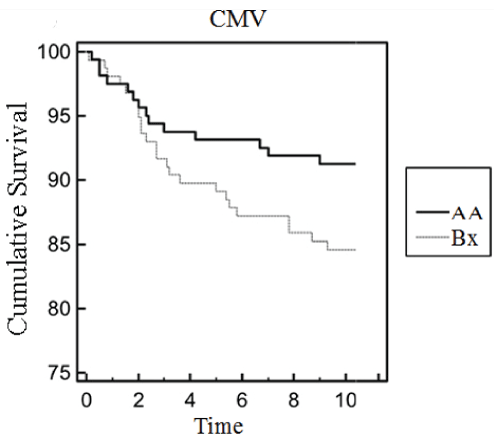 Figure 1: Survival Analysis.
Figure 1: Survival Analysis.
Cumulative incidence of cytomegalovirus infection in kidney transplant
recipients stratified by KIR AA and Bx haplotype.
View Figure 1
Discussion
Cytomegalovirus (CMV) is a significant cause of morbidity among patients receiving chronic maintenance immunosuppression and is often considered to show viral infection.[15] There exist well-defined risk factors for CMV disease, such as CMV seropositive-status of the donor and recipient, the immunosuppressive regimen, and the type of transplanted organ [16]. Cytomegalovirus (CMV) seropositivity is known to have a major impact on the repertoire of antigen specific T cells as an estimated 10% of circulating T cells is CMV-antigen specific in seropositive individuals [17]. In addition, the changes generally observed with an aged immune system, like a decreased CD4/CD8 ratio and expansion of CD28 negative CD8 positive T cells seem to be related to CMV sero-positivity [18,19]. In our study, all of the donors and recipients undergo live related transplantation were CMV IgG sero-positive. This was due to the higher incidence of CMV infection among Indian population [20]. We were not able to differentiate between CMV Donor-ve/Recipient-ve, Donor-ve/Recipient+ve, Donor+ve/Recipient-ve categories due to lack of seroreactive positive cases.
Several infectious diseases have been confirmed to be associated with certain KIR receptor/HLA ligand combinations [21]. Human NK cells that have matured in the presence of HLA ligands for KIR receptors developed more potent cytotoxic capacity than those maturing in the absence of such ligands through a mechanism called 'licensing' [22]. Therefore, in vitro at least, the lysis of CMV-infected cells by NK cells could be more complex than a simple missing KIR ligand mechanism [23]. It has been suggested that inflammatory signals induced by infection could break NK-cell tolerance and even in the absence of the HLA ligand, unlicensed NK cells could be activated to kill the virus-infected cells. The contribution of non-KIR family receptors could also become predominant in the immune response of NK cells to viral infections.
In the present study inhibitory receptor KIR2DL1 showed protective effect for CMV infected renal transplant cases. The combinatorial analysis revealed significant protective associations against KIR2DL2-HLAC1, KIR2DL3-HLA-C1 and KIR3DL1-HLA-Bw4 combinations for CMV infected cases. KIR, the best characterized group of NK receptors are allotype and isotype specific. KIR and HLA are present on different chromosomes and therefore are differently inherited. NK-cell activity is probably a result of the balance between activating and inhibitory receptors [24].
CMV causes a latent infection in majority of infected individuals. In immune-compromised individuals it can reactivate, and cause a life threatening illness. Earlier reports have emphasized upon the importance of NK cells and their KIR receptor in controlling CMV reactivation after hematopoietic cell transplant (HCT). More specifically, donors with more than one activating KIR gene were associated with 65% reduction in CMV reactivation [25] and the same effect has been seen in kidney transplants [26]. In the HCT setting, it was determined that the number of activating KIR genes in the donors but not in the recipients, was associated with protection from CMV reactivation and that the protective effect was highest when the donor genotype contained >5 activating KIR genes or at least a combination of KIR2DS2 and KIR2DS4 [27]. Although the frequency of KIR2DS2 and 2DS4 is prominent in the present study among CMV infected individuals but it did not reveal any significance.
Studies on murine NK cells [28,29] have shown that if mature NK cells from wild-type are transferred into a host with a different MHC class I environment, the cells can be reprogrammed to their new environment. This may explain the increased expression of activating KIR after transplant reported in several earlier studies. Mature NK cells can be re-educated as their new environment dictates, the new environment being allo-transplantation and /or CMV reactivation. In the matched relative donor transplant typically do encode for the same HLA type in donor and recipient but, it is possible that minor histocompatibility antigens or other HLA (e.g. non classical HLA-G-E) could serve as ligand that could influence this enhancement of expression.
In lieu of the hypothesis that presence of activating KIR genes may downplay the incidence of CMV infection we have found prolonged graft survival among individuals possessing KIR Bx haplotype which possesses number of activating KIR genes. However, the question is what specific CMV-associated factors might influence KIR gene expression? KIR genes are known to be regulated by epigenetic factors [30,31], and it is possible that either viral or host factors have some effect. A cohort of 286 kidney allograft recipients in the present study have demonstrated evidence of protective effect of activating KIR on the graft survival rate among CMV infected cases. While our data is in concordance with recent evidence from hematopoietic stem cell transplant recipients, the major limitation of our results is that all the cases under this study were seropositive for CMV. Further the data for CMV recurrence is very less so Donor/Recipient combinations were not highly informative.
Acknowledgements
We acknowledge the financial support to Department of Biotechnology, New Delhi to carry out this work. The authors declare that there is no conflict of interest, either of a financial or other nature, in relation to this work.
References
-
Fishman JA, Emery V, Freeman R, Pascual M, Rostaing L, et al. (2007) Cytomegalovirus in transplantation - challenging the status quo. Clin Transplant 21: 149-158.
-
Fishman JA (2007) Infection in solid-organ transplant recipients. N Engl J Med 357: 2601-2614.
-
Casazza JP, Betts MR, Price DA, Precopio ML, Ruff LE, et al. (2006) Acquisition of direct antiviral effector functions by CMV-specific CD4+ T lymphocytes with cellular maturation. J Exp Med 203: 2865-2877.
-
Harari A, Zimmerli SC, Pantaleo G (2004) Cytomegalovirus (CMV)-specific cellular immune responses. Hum Immunol 65: 500-506.
-
Villard J (2006) Immunity after organ transplantation. Swiss Med Wkly 136: 71-77.
-
Wang H, Grzywacz B, Sukovich D, McCullar V, Cao Q, et al. (2007) The unexpected effect of cyclosporin A on CD56+CD16- and CD56+CD16+ natural killer cell subpopulations. Blood 110: 1530-1539.
-
Chiossone L, Vitale C, Cottalasso F, Moretti S, Azzarone B, et al. (2007) Molecular analysis of the methylprednisolone-mediated inhibition of NK-cell function: evidence for different susceptibility of IL-2- versus IL-15-activated NK cells. Blood 109: 3767-3775.
-
Martin MP, Gao X, Lee JH, Nelson GW, Detels R, et al. (2002) Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet 31: 429-434.
-
Shastry A, Sedimbi SK, Rajalingam R, Nikitina-Zake L, Rumba I, et al. (2008) Combination of KIR 2DL2 and HLA-C1 (Asn 80) confers susceptibility to type 1 diabetes in Latvians. Int J Immunogenet 35: 439-446.
-
Cook M, Briggs D, Craddock C, Mahendra P, Milligan D, et al. (2006) Donor KIR genotype has a major influence on the rate of cytomegalovirus reactivation following T-cell replete stem cell transplantation. Blood 107: 1230-1232.
-
Chen C, Busson M, Rocha V, Appert ML, Lepage V, et al. (2006) Activating KIR genes are associated with CMV reactivation and survival after non-T-cell depleted HLA-identical sibling bone marrow transplantation for malignant disorders. Bone Marrow Transplant 38: 437-444.
-
Gumá M, Budt M, Sáez A, Brckalo T, Hengel H, et al. (2006) Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood 107: 3624-3631.
-
Prakash S, Alam S, Sharma RK, Sonawane A, Imran M, et al. (2013) Distribution of Killer cell immunoglobulin like receptor genes in end stage renal disease among North Indian population. Hum Immunol 74: 1339-1345.
-
Kreijveld E, van der Meer A, Tijssen HJ, Hilbrands LB, Joosten I (2007) KIR gene and KIR ligand analysis to predict graft rejection after renal transplantation. Transplantation 84: 1045-1051.
-
Murray BM, Amsterdam D, Gray V, Myers J, Gerbasi J, et al. (1997) Monitoring and diagnosis of cytomegalovirus infection in renal transplantation. J Am Soc Nephrol 8: 1448-1457.
-
Bhadauria D, Sharma RK, Kaul A, Prasad N, Gupta A, et al. (2012) Cytomegalovirus disease in renal transplant recipients: a single-center experience. Indian J Microbiol 52: 510-515.
-
Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, et al. (2005) Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med 202: 673-685.
-
Wikby A, Johansson B, Olsson J, Löfgren S, Nilsson BO, et al. (2002) Expansions of peripheral blood CD8 T-lymphocyte subpopulations and an association with cytomegalovirus seropositivity in the elderly: the Swedish NONA immune study. Exp Gerontol 37: 445-453.
-
Derhovanessian E, Larbi A, Pawelec G (2009) Biomarkers of human immunosenescence: impact of Cytomegalovirus infection. Curr Opin Immunol 21: 440-445.
-
Chakravarti A, Kashyap B, Matlani M (2009) Cytomegalovirus infection: an Indian perspective. Indian J Med Microbiol 27: 3-11.
-
Ahlenstiel G, Martin MP, Gao X, Carrington M, Rehermann B (2008) Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. J Clin Invest 118: 1017-1026.
-
Anfossi N, André P, Guia S, Falk CS, Roetynck S, et al. (2006) Human NK cell education by inhibitory receptors for MHC class I. Immunity 25: 331-342.
-
Carr WH, Little AM, Mocarski E, Parham P (2002) NK cell-mediated lysis of autologous HCMV-infected skin fibroblasts is highly variable among NK cell clones and polyclonal NK cell lines. Clin Immunol 105: 126-140.
-
Lanier LL (2001) On guard--activating NK cell receptors. Nat Immunol 2: 23-27.
-
Chen C, Busson M, Rocha V, Appert ML, Lepage V, et al. (2006) Activating KIR genes are associated with CMV reactivation and survival after non-T-cell depleted HLA-identical sibling bone marrow transplantation for malignant disorders. Bone Marrow Transplant 38: 437-444.
-
Stern M, Elsässer H, Hönger G, Steiger J, Schaub S, et al. (2008) The number of activating KIR genes inversely correlates with the rate of CMV infection/reactivation in kidney transplant recipients. Am J Transplant 8: 1312-1317.
-
Zaia JA, Sun JY, Gallez-Hawkins GM, Thao L, Oki A, et al. (2009) The effect of single and combined activating killer immunoglobulin-like receptor genotypes on cytomegalovirus infection and immunity after hematopoietic cell transplantation. Biol Blood Marrow Transplant 15: 315-325.
-
Joncker NT, Shifrin N, Delebecque F, Raulet DH (2010) Mature natural killer cells reset their responsiveness when exposed to an altered MHC environment. J Exp Med 207: 2065-2072.
-
Elliott JM, Wahle JA, Yokoyama WM (2010) MHC class I-deficient natural killer cells acquire a licensed phenotype after transfer into an MHC class I-sufficient environment. J Exp Med 207: 2073-2079.
-
Li G, Yu M, Weyand CM, Goronzy JJ (2009) Epigenetic regulation of killer immunoglobulin-like receptor expression in T cells. Blood 114: 3422-3430.
-
Chan HW, Miller JS, Moore MB, Lutz CT (2005) Epigenetic control of highly homologous killer Ig-like receptor gene alleles. J Immunol 175: 5966-5974.





