Journal of Genetics and Genome Research
Understanding the Host Epigenetics in Mycobacterium tuberculosis Infection
Vinod Yadav1, Ved Prakash Dwivedi1, Debapriya Bhattacharya1, Ashwani Mittal2, Prashini Moodley1 and Gobardhan Das1,3*
1School of Laboratory Medicine and Medical Science, College of Health Sciences, Nelson R Mandela School of Medicine, University of KwaZulu-Natal, South Africa
2Department of Biochemistry, University College, Kurukshetra University, India
3Special Centre for Molecular Medicine (SCMM), Jawaharlal Nehru University, India
*Corresponding author:
Prof. Gobardhan Das, School of Laboratory Medicine and Medical Science, Nelson Mandela Medical School, University of KwaZulu-Natal, Durban-4001, South Africa
&
Special Center for Molecular Medicine (SCMM), Jawaharlal Nehru University, New Delhi-110067, India, E-mail: gobardhan.das07@gmail.com
J Genet Genome Res, JGGR-2-016, (Volume 2, Issue 1), Research Article; ISSN: 2378-3648
Received: April 15, 2015 | Accepted: June 24, 2015 | Published: June 27, 2015
Citation: Yadav V, Dwivedi VP, Bhattacharya D, Mittal A, Moodley P, et al. (2015) Understanding the Host Epigenetics in Mycobacterium tuberculosis Infection. J Genet Genome Res 2:016. 10.23937/2378-3648/1410016
Copyright: © 2015 Yadav V, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Epigenetics denotes to study the heritable changes occurred in the gene function without any changes in DNA sequence. These epigenetic changes are known to be governed by various factors viz. stress, infection, nutrients, drugs and toxicological agents etc. Recently, it has been identified that different microorganisms can cause the epigenetic changes in host. In this review we intend to address about the epigenetic changes occurred in host by Mycobacterium tuberculosis (M.tb) infection and then elaborate the current state of research about how Mtb. modulates host epigenome. M.tb induced epigenetic modifications which either leads to promote host defense or M.tb survival. Therefore, M.tb can be considered as potential modulator of host epigenome and consequently, these epigenetic changes can be beneficial or disastrous to M.tb. Currently, there is huge advances in sequencing technology and this can lead to a better understanding of the roles of epigenetics in the tuberculosis and other infectious diseases. Subsequently, therapeutic targeting of the epigenome can be potentially helpful in treatment of Mtb infection.
Keywords
Mycobacterium tuberculosis (M.tb), Epigenetics, Macrophage, Chromatin modification
Introduction
It was in 1942 when C.H. Waddington first coined the term epigenetics. According to him there is no direct relationship between a gene and its phenotype. He considered epigenetics as a part of development biology and in his opinion many times genotype and phenotype variations are not associated and phenotype differences do not necessarily involve change in genotype [1]. Since 40s epigenetics became the topic of interest among the scientific community. Now everybody wants to know that what goes on beyond the DNA and majority says that only epigenetics can address this question. In simple words, epigenetics can be defined as the science of stable and heritable changes occurred in cells without the change in DNA sequence for example induction, repression or silencing of gene expression. In addition, these epigenetic mechanisms are responsible for chromatin dynamics and which can further regulate diverse cellular processes like DNA repair, recombination and gene expression (Figure 1). It is interesting to note that sometimes these epigenetic changes can stably inherit through cell divisions. Furthermore, these changes can be erased or modified during differentiation, development and under different environmental stimuli. Such changes are mediated by various modifications on DNA and its associated histones. These modifications include methylation, acetylation, phosphylation and ubquitylation and all of these associated with chromatin dynamics and organization [2-10]. However all these modifications are not fulfilling the definition of epigenetics in terms of heritability? To follow this definition these modifications must be transmitted during the DNA replication [11,12]. Additionally, all these epigenetic processes play decisive role in development of cancer, where repression of tumor suppressor gene takes place. Also, various pathogens modulate these epigenetic processes to tackle host immune response and causes chronic illnesses [13-16]. Several line of evidences have established the role of pathogenic microbe on modulation of host transcriptional program by enforce changes in key cellular processes genes like genes responsible for immunity, apoptosis and survival etc. The fact is well established that most of the successful human pathogenic microorganisms were evolved in such a manner that they can effectively overcome against the host defense [17-22]. These successful human pathogenic microorganisms are able to hijack and reprogram host genome by targeting important cellular signaling pathways and transcription factors. For instance, many bacterial products can modify, activate, inhibit or degrade transcription factors and other key cellular proteins [18,21,23]. One such example of successful human pathogenic microorganisms is M.tb causative agent of Tuberculosis (TB) (Figure 1).
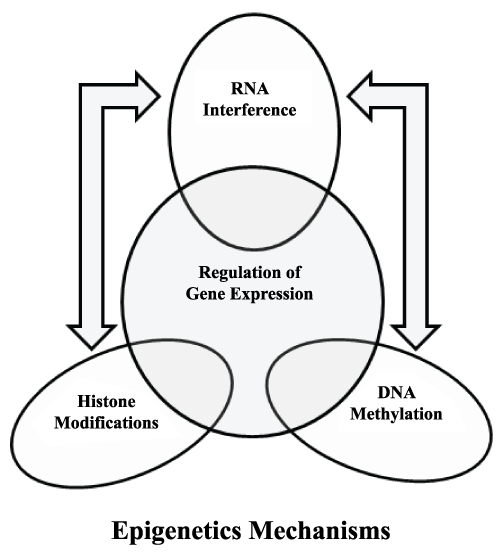
Figure 1: Various epigenetic mechanisms plays role in the regulation of gene expression
View Figure 1
TB, a life threatening infectious diseases caused by the obligatory, aerobic bacillus M.tb is becoming a major global health problem [24]. It is a communicable disease transmitted via droplets produced through coughing and sneezing by individual having active tuberculosis. M.tb infection in healthy individual often remains latent or asymptomatic whereas active disease appears in the individual having suboptimal immune functioning. However, in latent infection M.tb persists in dormant condition which can subsequently cause active tuberculosis in immune compromised individual. According to WHO recommendations M.tb infected individuals can be treated using four first line drugs: isoniazid, rifampicin, ethambutol and pyrazinamide for six months also known as ATT (Anti-Tuberculosis Therapy). However, therapeutic utility of these drugs is now jeopardized by emergence of MDR-M.tb, XDR-M.tb and TDR-M.tb strains [25-29].
Whereas treatment of MDR-TB requires more expensive and toxic drugs for about twenty months and which has much lower success rate. Recent report released by WHO estimated 450 thousand people developed MDR-TB and 30% of which died due to tuberculosis. Additionally, it was observed that out of total MDR-TB cases about 10% develops into XDR-TB.
Globally, about one third of total population is infected with M.tb but out of that ~10% of these individuals will diagnosed for active TB. This is an important question to address that why not the all individual infected with M.tb develop clinical disease. After the inhalation of M.tb, human body can respond in three different ways either this infection will leads to active TB or human immune response clears TB bug from system or M.tb will undergo latency. Latent TB is hard to detect and it can relapse and cause active disease in different immune-compromised state of human. Altogether TB progression mainly depends on the ability of host immune response against M.tb. [30]. This immune response is affected by either internal factor as host genetic makeup or external factors which affect epigenetics of host like environment, nutrition and stress etc.
In this review, we contend that how M.tb modulates host epigenome to overcome the host defense and use this strategy to fight for its survival inside the host. After an overview about the epigenetic mechanisms governing chromatin dynamics, we explained that how these epigenetic mechanisms are targeted by M.tb to enforce change in host transcriptional program for its survival and persistence.
Types of Epigenetic Modifications
Eukaryotic chromatin is a complex structure which comprises of DNA and histone proteins and to fit into the tight space of nucleus it organizes itself into a dynamic higher order structure [31]. Organization of these structures plays an important role in various nuclear processes like DNA replication, transcription, and recombination and DNA repair [5,32].
Several lines of investigations have suggested the presence of different level of chromatin organization. Starting from nucleosomes which consist of core histone octamers (H2A, H2B, H3 and H4) circled by 147 base pair DNA and linked to other nucleosome via linker histone (H1). In next level of chromatin organization the DNA complexed with histone forms a chromatin fiber (30nm) and which further condensed into compact chromatin. These higher order chromatin structure either forms euchromatin (transcriptionally active) or hetrochromatin (transcriptionally inactive). Formation of euchromatin and hetrochromatin is a dynamic process and tightly regulated by various remodeling and modifying mechanisms [33-36]. Altogether, chromatin structure is vitally responsible for various kind of nuclear processes outcome like gene expression, silencing, DNA replication, transcription, recombination, DNA repair and genome stability [37-40]. Histone modification acts as important epigenetic marks. Histone octamer present in nucleosomes can undergo various covalent additions of different chemical groups and called as post translational modifications (PTMs) of histone [41-46].
In addition to modifications of histone proteins, DNA methylation is a chemical modification occurs when C5 position of cytosine in CpG-rich regions of DNA (CpG island) is changed due to transfer of methyl group by DNA methytransferase (DNMTs) either to establish methylation (DNMT3a and DNMT3b) or to copy methylation pattern (DNMT1) to newly synthesized DNA during replication. DNA methylation is reversible by two independent mechanisms either passively during DNA replication when 5mC (5 methyl Cytosine) is not copied or actively in DNA repair process and demethylation during chemical modification of 5mC [47-49].
Recently, whole mammal genome and transcriptome has been sequenced and it was observed that huge number of transcribed RNA is not translated into the protein which suggest about the presence of non-coding RNA (nc RNA) in the transcribed RNAs and these nc RNA plays a pivotal regulatory role in various cellular functions similar to histone modifications and DNA methylation. In 2001 nc RNA were identified which is also known as microRNA (miRNA) and compose a large family of small non-coding RNA viz., piwi-interacting RNA (piRNA) , small interfering RNA (siRNA), small nucleolar RNA (snoRNA) and small nuclear RNA (snRNA). These miRNA target mRNA at a translational level by repressing gene expression and act as endogenous gene silencers. The genome of animals, plants, and viruses contains highly conserved miRNA. Recent reports suggest that miRNA might play a pivotal function in regulating aboutone third of mammalian genes [50-59].
M.tb Infection Alters Histone Modifications
Several studies have shown that certain infectious agents like Helicobacter pylori, Streptococcus bovis, Chlamydia pneumoniae, Campylobacter rectus, Epstein-Barr virus, hepatitis viruses, Human papilloma virus, polyomaviruses, etc. can contribute to the host epigenetic changes resulting in the onset and progression of some diseases, especially in malignancies. However these epigenetic modifications induced by these infectious agents in host cells are largely ill defined. Possibly, these infectious agents like viruses, bacteria and other parasitic microorganisms have a lot of complex epigenetic regulatory mechanisms, which may cause epigenetic deregulation in their respective hosts. As an obligate intracellular pathogen, M.tb has also developed numerous mechanisms of hijacking cellular processes to tackle against the host immune response. M.tb which also causes latent infection is undoubtly taking advantage from the epigenetic changes occurred in host after its infection. These changes make the M.tb friendly environment inside the host cells and favor its survival, growth and latency (Figure 2,3).
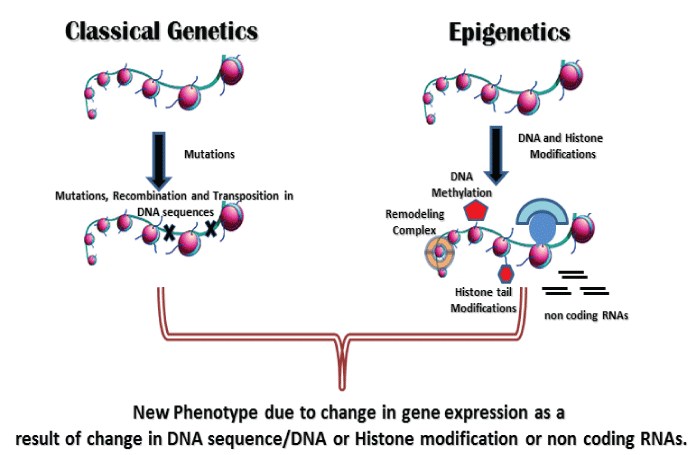
Figure 2: Emergence of new phenotype due to change in DNA sequence (Genetics) or DNA/Histone modification/non coding RNAs (Epigenetics).
View Figure 2
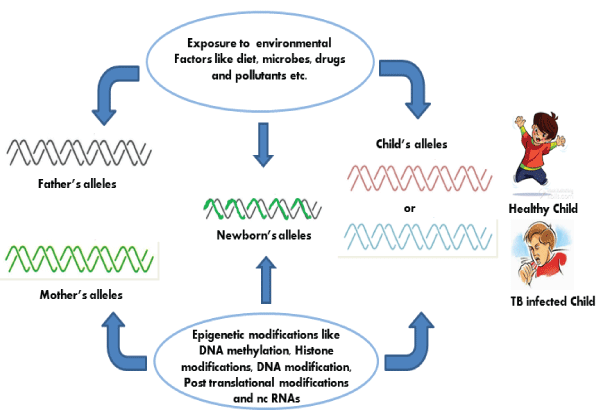
Figure 3: Along with host genetics, epigenetics plays an important role in Tuberculosis.
View Figure 3
Histone modifications are divided into eight different classes of post-translational modification which have about 60 distinct modification sites viz. proline isomerization, arginine deimination, serine,glutamate poly-ADP ribosylation, lysine and arginine methylation, threonine and thyrosine phosphorylation, ubiquitination and sumolylation, and lysine acetylation and altogether cause the differences in dynamics and function of chromatin [5,41]. For instance, active chromatin is linked with the PTM in histone 3 lysine 4 residue trimethylation (H3K4me3) whereas histone 3 lysine 9 residue trimethylation (H3K9me3) is the representative of repressed chromatin [42,43]. All these covalent modifications of chromatin are guided by different enzymes. Some of these enzymes act as writers like kinases, histone methyltransferase (HMTs) and histone acetyltransferase (HATs) and others functions as erasers like phosphatase, histone demethylase (HDMs), histone deacetylase (HDACs) [44-46]. Histone tail acetylation by HATs is usually linked with activation of chromatin by increases the space between nucleosomes whereas deacetylation via HDACs leads to suppression of gene expression [40]. Both the epigenetic mechanism of writing and erasing on histones are indispensable for the proper regulation of gene expression and absolute homeostasis is required for these processes. Altogether these histone codes for gene regulation are complex in nature and works in synergy with various other epigenetic mechanisms which ultimately determine the outcome of gene activity. Furthermore, it will be important to look about the genome wide modification in histone codes after M.tb infection in the host cells. This process of maintaining the histone code is so beautifully followed by the cells that the same stimuli can induce writing on histone tails at one locus whereas erasers removes some residues from histone tails. Therefore, it will be worth to address that what governs histone modifying enzymes to such a precision so that they can modify a set of specific gene loci out of whole genome of cell (Figure 2).
To combat the attack of M.tb infection, natural killer cells and activated T cells secrete lot IFN- which induces the major histocompatibility complex class II (MHC class II) expression in numerous cell kinds [60-63]. Interaction of IFN- with its cognate receptor on cell surface activates Janus tyrosine kinase-signal transducer and activator of transcription (JAK -STAT1) signaling cascade and regulates the transcription of several genes including CIITA trans-activator. This trans-activator plays pivotal role in the transcription of the MHC class II complex genes [64]. However, M.tb is able to inhibit some of genes like HLA-DR, CD64 and CIITA which are induced by IFN- , despite activation of normal JAK -STAT1 signaling cascade. Reports suggest that M.tb is modulating histone modifications and chromatin remodeling at specific promoter to inhibit the transcription of these immune genes [24,65]. The blocked of CIITA gene transcription is due to the histone deacetylation at the CIITA promoter and inhibition of SWI/SNF binding and conversely M.tb induces the binding of the transcriptional repressor C/EBP to CIITA promoter However the detailed mechanism for this process is not given but the author hypothesized that 19KDa protein induced prolonged TLR2 signaling can be responsible for the expression/activation/repression of some identified transcription factors therefore additional studies are required to gain further insights [63,66]. Similarly HLA-DR expression was inhibited in M.tb infected cells by impairing the histone acetylation, and recruitment of corepressor at the HLA-DR promoter [67]. Additional mechanisms are required to be determined to exactly understand that how M.tb induces the repressor recruitment to these promoter in a specific way.
A recent report concludes that optimal immune activation by CDC1551 (M.tb sensitive strain) is sufficient to clear it form host, however HN878 (M.tb resistant strain) can sub optimally induce immune activation and over expression of gene responsible for the host lipid metabolism and ultimately enables better intracellular survival of HN878 bacilli. From this study, it is oblivious that both strains of M.tb are behaving differentially and inducing or inhibiting different set of genes whereas the precise molecular details about these changes are still unclear. Further studies are needed to completely decipher about the epigenetic changes happening in host and M.tb which is making one strain sensitive and other as resistant [68].
M.tb Infection Modulates Non Coding RNAs Expression
The miRNAs have been shown to play decisive roles in development, differentiation, apoptosis, and oncogenesis [53,54]. Synthesis of miRNA occurs in nucleus as long primary transcripts (pri-miRNA) and has imperfect hairpin structures. Transcription of miRNA is majority mediated by RNA polymerase II (Pol II), however some are also transcribed by Pol III. Then with help of RNase III, Drosha and DGCR8 process this pri-miRNA into precursor miRNA (pre-miRNA). Subsequently, the RNAse III Dicer enzyme cooperates with other proteins and processes the pre-miRNA into a mature miRNA as well as a complementary fragment called as miRNA. This mature miRNA in combination of RNA-induced silencing complex (RISC) governs gene regulation processes [55-59]. Number of lines of investigation describes that miRNA are involved in the regulation of important cellular pathways, such as proliferation, cell death, angiogenesis, invasion and in chromatin structure dynamics and genome organization in nucleus which has added additional complexity in epigenetic mechanisms [60-62] (Figure 2).
MicroRNAs (miRNAs) are non-coding single stranded RNAs of approximately 22 nucleotides in length which are important component of epigenetic mechanisms and can specifically modulate gene expression. Considering the central role of miRNAs in development and disease, the authors in recent report measured by the levels of miRNA in the blood of TB patients.
In above mentioned study it was observed that, in comparison of control individual's serum TB patient's serum has 59 overexpressed miRNAs whereas 33 miRNAs were under-expressed which significantly concludes that miRNAs are playing important role in active TB. Authors hypothesized that the serum miRNAs can potentially be harvested as the novel biomarker for the diagnosis and evaluation of the status of TB [63]. A recent study describes about the role of miR115 in macrophages infected with BCG. The expression of miR115 is unregulated in macrophages infected with BCG and it reprograms diverse cellular signaling cascades in macrophages and implicates important role in pathogenesis and survival of tuberculosis [67].Interestingly, human macrophages after mycobacterial infection induce a specific modulation of miRNAs in macrophages [69]. In addition, BCG down-regulates miR-29 expression in, CD4+ T cells, CD8+ T cells and natural killer cells [70]. Altogether these findings opens a new possibility in illuminating the connection between M.tb infection and role of miRNAs, and further research is needed to find out more about this association.
M.tb Infection Modifies DNA Methylation
DNA methylation has huge role in various cellular processes like differentiation, development, reprogramming, gene silencing and induction of various diseases including cancer [49]. Methylation at CpG causes silencing effects on gene expression by preventing association of various transcription or DNA binding factors to their consensus binding sites present in CpG islands or by recruit co-repressor to methylated CpG nucleotides and modify chromatin into repressive form. Majorly, methylation of DNA at enhancers or promoter region of gene causes transcriptional repression or silencing. However, in comparison to histone modification DNA methylation is quite stable epigenetic change and very hard to reversible and this physiological epigenetic process may lead to long term silencing of gene expression [48,49] (Figure 2).
In a recent finding it was observed that BCG mediated epigenetic reprogramming of innate immune cells confers the absolute survival in all BCG-vaccinated SCID mice in comparison of only one third survival in control SCID mice. The BCG vaccine is able to do so in mononuclear phagocytes via NOD2-mediated epigenetic change at the level of H3K4me3. In this row further insights are needed for understanding the epigenetic reprogramming of innate immunity during M.tb infection and use this understanding for designing the effective vaccine and adjuvants which can selectively induce epigenetic modifications to ultimately help to generate the effective memory response against M.tb [71].
Notably, recent studies have reported the dynamic nature of DNA methylation in dendritic cells (DCs) upon infection of M.tb. Methylation profiles of DCs before and after M.tb infection showed significant differentially methylated regions (DMRs) in single-base pair resolution analyzed data. Further analysis showed that changes in methylation were largely observed at low CpG density regions and distal regulatory regions or enhancers rather in high CpG density regions and gene promoters. Interestingly, about one third of the genes with changed expression after M.tb infection have promoter or close proximity DMR. Altogether this report suggested the role of DNA methylation in M.tb infection [72].
Another study using human macrophages suggested the modulation in DNA methylation status after M.tb infection. Using Agilent Human CpG Island Microarray, authors found significant alterations in methylation levels of inflammatory genes in M.tb infected human macrophages. Substantial increase in methylation was observed at the promoter region of interleukin-17 receptor gene in comparison with other IL-17 family members and receptors in macrophages. Additionally, the change in methylation patterns in host depends on type of M.tb strain and host genotype. Authors also concluded that these signature DNA methylation profiles may be exploited to use for the diagnosis of M.tb infection clinically [73].
M.tb induced changes in methylation of histone and acetylation in histone is evident from a report suggesting the role of M.tb 6kDa, ESAT6 (early secreted antigen 6) in reduction of IFN-γ- induced histone H3K4 methylation and CIITA pI locus histone acetylation in M.tb infected macrophages. In addition this report suggests that inhibition of type IV and type I CIITA expression is mediated by ESAT6 via different signaling pathways. Inhibited expression of type IV CIITA was found to be Toll-like receptor-2 (TLR2) dependent, whereas type I CIITA expression inhibitionis TLR2independent. In conclusion, both of these mechanisms have capacity to do chromatin remodeling and induce the differential effects [74]. M.tb secreted Enhanced intracellular survival (Eis) protein plays important role in increased survival of Mycobacterium smegmatisin macrophages by acetylating the histone proteins [75]. Recently, Siddle et al. has assessed change in miRNA profile in human DCs infected with M.tb. Approximately, 40% of miRNAs were differentially expressed in miRNAs in M.tb infected DCs during analysis of genome wide expression profiling. The study further emphasized about the global role of miRNAs in M.tb infected cells and opens a new field in epigenetic research about the role of miRNAs in patho-physiology of TB. Furthermore additional studies are required to find out the detailed contributions of miRNAs in TB immunity, infection, disease and latency [76].
Different ethnic groups carry variations in the methylation pattern and status in the vitamin D receptor gene promoter CpG islands upon M.tb infection. Therefore, it can be extrapolated that M.tb susceptibility depends on the level of global methylation status and subsequently can be used for designing some strategies for early diagnosis of TB susceptibility in individuals [77]. Recent work explained that pathological over-expression of Matrix Metalloproteinases 1 (MMP-1) in TB is contributed by epigenetic mechanisms via histone acetylation. The report suggested that using inhibition of either HDAC or HAT enzymes blocked the M.tb induced MMP-1 expression in infected macrophages. Furthermore, M.tb infection reduced the expression of Class I HDACs whereas HDAC1 and 3expressions remained unaltered [78].
Future Directions
Human hosts are co-evolved with M.tb, which has given this bacterium the inimitable ability to exploit and survive in the human host. M.tb has unique strategies to modulate epigenetic mediators and repress host immune response genes. Recent research about M.tb induced epigenetic regulation in host opened new avenues on host M.tb epigenetics. Further understanding the function of epigenetics and its regulators in patho-physiology of tuberculosis is a challenging task. However determining the different epigenetic marks in chromatin after M.tb infection in various infected cells will aid us to unravel diverse mechanisms and strategies of M.tb for its survival. These investigations can provide us the opportunity to design therapeutic applications against TB. Furthermore, we can find some therapeutics agents which can revert different epigenetic processes and prevent active and latent TB. Altogether, this will open a new avenue in the research and development field of M.tb pathogenesis and epigenetic regulation. May be some day we can challenge M.tb in same way how it is challenging us today.
Acknowledgements
We would like to thank the University of KwaZulu-Natal, Durban, South Africa for the funding.
References
-
Waddington CH (2012) The epigenotype. 1942. Int J Epidemiol 41: 10-13.
-
Mehler MF (2008) Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog Neurobiol 86: 305-341.
-
Robertson KD (2005) DNA methylation and human disease. Nat Rev Genet 6: 597-610.
-
Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074-1080.
-
Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693-705.
-
Szyf M (2009) Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol 49: 243-263.
-
Cairns BR (2009) The logic of chromatin architecture and remodelling at promoters. Nature 461: 193-198.
-
Schoeftner S, Blasco MA (2009) A 'higher order' of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J 28: 2323-2336.
-
Payer B, Lee JT (2008) X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet 42: 733-772.
-
Boehm U, Klamp T, Groot M, Howard JC (1997) Cellular responses to interferon-gamma. Annu Rev Immunol 15: 749-795.
-
Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A (2009) An operational definition of epigenetics. Genes Dev 23: 781-783.
-
Feinberg AP (2007) Phenotypic plasticity and the epigenetics of human disease. Nature 447: 433-440.
-
Bracken AP, Helin K (2009) Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer 9: 773-784.
-
Esteller M (2007) Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 8: 286-298.
-
Gieni RS, Hendzel MJ (2009) Polycomb group protein gene silencing, non-coding RNA, stem cells, and cancer. Biochem Cell Biol 87: 711-746.
-
Flanagan JM (2007) Host epigenetic modifications by oncogenic viruses. Br J Cancer 96: 183-188.
-
Lefèvre T, Lebarbenchon C, Gauthier-Clerc M, Missé D, Poulin R, et al. (2009) The ecological significance of manipulative parasites. Trends Ecol Evol 24: 41-48.
-
Bhavsar AP, Guttman JA, Finlay BB (2007) Manipulation of host-cell pathways by bacterial pathogens. Nature 449: 827-834.
-
Hamon MA, Cossart P (2008) Histone modifications and chromatin remodeling during bacterial infections. Cell Host Microbe 4: 100-109.
-
Paschos K, Allday MJ (2010) Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends Microbiol 18: 439-447.
-
Ribet D, Cossart P (2010) Post-translational modifications in host cells during bacterial infection. FEBS Lett 584: 2748-2758.
-
Kincaid EZ, Ernst JD (2003) Mycobacterium tuberculosis exerts gene-selective inhibition of transcriptional responses to IFN-gamma without inhibiting STAT1 function. J Immunol 171: 2042-2049.
-
Pai RK, Pennini ME, Tobian AA, Canaday DH, Boom WH, et al. (2004) Prolonged toll-like receptor signaling by Mycobacterium tuberculosis and its 19-kilodalton lipoprotein inhibits gamma interferon-induced regulation of selected genes in macrophages. Infect Immun 72: 6603-6614.
-
Pattenden SG, Klose R, Karaskov E, Bremner R (2002) Interferon-gamma-induced chromatin remodeling at the CIITA locus is BRG1 dependent. EMBO J 21: 1978-1986.
-
Almeida Da Silva PE, Palomino JC (2011) Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother 66: 1417-1430.
-
Davies J (1996) Origins and evolution of antibiotic resistance. Microbiologia 12: 9-16.
-
Gillespie SH (2002) Evolution of drug resistance in Mycobacterium tuberculosis: clinical and molecular perspective. Antimicrob Agents Chemother 46: 267-274.
-
Shah NS, Wright A, Bai GH, Barrera L, Boulahbal F, et al. (2007) Worldwide emergence of extensively drug-resistant tuberculosis. Emerg Infect Dis 13: 380-387.
-
Velayati AA, Farnia P, Masjedi MR, Ibrahim TA, Tabarsi P, et al. (2009) Totally drug-resistant tuberculosis strains: evidence of adaptation at the cellular level. Eur Respir J 34: 1202-1203.
-
Flynn JL, Chan J (2001) Immunology of tuberculosis. Annu Rev Immunol 19: 93-129.
-
Woodcock CL, Dimitrov S (2001) Higher-order structure of chromatin and chromosomes. Curr Opin Genet Dev 11: 130-135.
-
Kretsovali A, Agalioti T, Spilianakis C, Tzortzakaki E, Merika M, et al. (1998) Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol Cell Biol 18: 6777-6783.
-
Kornberg RD, Lorch Y (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98: 285-294.
-
Oudet P, Gross-Bellard M, Chambon P (1975) Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell 4: 281-300.
-
Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251-260.
-
Robinson PJ, Fairall L, Huynh VA, Rhodes D (2006) EM measurements define the dimensions of the "30-nm" chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci U S A 103: 6506-6511.
-
Cosgrove MS, Boeke JD, Wolberger C (2004) Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol 11: 1037-1043.
-
Goll MG, Bestor TH (2002) Histone modification and replacement in chromatin activation. Genes Dev 16: 1739-1742.
-
Grant PA (2001) A tale of histone modifications. Genome Biol 2: REVIEWS0003.
-
Shahbazian MD, Grunstein M (2007) Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem 76: 75-100.
-
Peterson CL, Laniel MA (2004) Histones and histone modifications. Curr Biol 14: R546-551.
-
Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, et al. (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86-90.
-
Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, et al. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120-124.
-
Zhou Y, Kim J, Yuan X, Braun T (2011) Epigenetic modifications of stem cells: a paradigm for the control of cardiac progenitor cells. Circ Res 109: 1067-1081.
-
Bhutani N, Burns DM, Blau HM (2011) DNA demethylation dynamics. Cell 146: 866-872.
-
Chen ZX, Riggs AD (2011) DNA methylation and demethylation in mammals. J Biol Chem 286: 18347-18353.
-
Fatemi M, Hermann A, Gowher H, Jeltsch A (2002) Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur J Biochem 269: 4981-4984.
-
Siedlecki P, Zielenkiewicz P (2006) Mammalian DNA methyltransferases. Acta Biochim Pol 53: 245-256.
-
Deaton AM, Bird A (2011) CpG islands and the regulation of transcription. Genes Dev 25: 1010-1022.
-
Sontheimer EJ, Carthew RW (2005) Silence from within: endogenous siRNAs and miRNAs. Cell 122: 9-12.
-
Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, et al. (2005) Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37: 766-770.
-
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15-20.
-
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, et al. (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 101: 2999-3004.
-
Ambros V (2004) The functions of animal microRNAs. Nature 431: 350-355.
-
Cai X1, Hagedorn CH, Cullen BR (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10: 1957-1966.
-
Lee Y, Kim M, Han J, Yeom KH, Lee S, et al. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051-4060.
-
Borchert GM, Lanier W, Davidson BL (2006) RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 13: 1097-1101.
-
Lee Y, Jeon K, Lee JT, Kim S, Kim VN (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21: 4663-4670.
-
Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858-862.
-
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281-297.
-
Cheng AM, Byrom MW, Shelton J, Ford LP (2005) Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 33: 1290-1297.
-
Hua Z, Lv Q, Ye W, Wong CK, Cai G, et al. (2006) MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One 1: e116.
-
Pennini ME, Liu Y, Yang J, Croniger CM, Boom WH, et al. (2007) CCAAT/enhancer-binding protein beta and delta binding to CIITA promoters is associated with the inhibition of CIITA expression in response to Mycobacterium tuberculosis 19-kDa lipoprotein. J Immunol 79: 6910-6918.
-
Fu Y, Yi Z, Wu X, Li J, Xu F (2011) Circulating microRNAs in patients with active pulmonary tuberculosis. J Clin Microbiol 49: 4246-4251.
-
Ghorpade DS, Leyland R, Kurowska-Stolarska M, Patil SA, Balaji KN (2012) MicroRNA-155 is required for Mycobacterium bovis BCG-mediated apoptosis of macrophages. Mol Cell Biol 32: 2239-2253.
-
Pennini ME, Pai RK, Schultz DC, Boom WH, Harding CV (2006) Mycobacterium tuberculosis 19-kDa lipoprotein inhibits IFN-gamma-induced chromatin remodeling of MHC2TA by TLR2 and MAPK signaling. J Immunol 176: 4323-4330.
-
Wang Y, Curry HM, Zwilling BS, Lafuse WP (2005) Mycobacteria inhibition of IFN-gamma induced HLA-DR gene expression by up-regulating histone deacetylation at the promoter region in human THP-1 monocytic cells. J Immunol 174: 5687-5694.
-
Koo MS, Subbian S, Kaplan G (2012) Strain specific transcriptional response in Mycobacterium tuberculosis infected macrophages. Cell Commun Signal 10: 2.
-
Sharbati J, Lewin A, Kutz-Lohroff B, Kamal E, Einspanier R, et al. (2011) Integrated microRNA-mRNA-analysis of human monocyte derived macrophages upon Mycobacterium avium subsp. hominissuis infection. PLoS One 6: e20258.
-
Ma F, Xu S, Liu X, Zhang Q, Xu X, et al. (2011) The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol 12: 861-869.
-
Kleinnijenhuis J, Quintin J, Preijers F, Joosten LA, Ifrim DC, et al. (2012) Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc Natl Acad Sci U S A 109: 17537-17542.
-
Barreiro LB, Pacis A, Tailleaux L, Yotova V, Grenier JC, et al. (2013) Widespread changes in DNA methylation at CpG island shores and distal regulatory regions in response to a bacterial infection. In ashg.
-
IP M, Zheng L, Leung ETY, Lee N, Lui G, et al. (2014) Unraveling Methylation Changes of Host Macrophages in Mycobacterium tuberculosis Infection. In asm
-
Kumar P, Agarwal R, Siddiqui I, Vora H, Das G, et al. (2012) ESAT6 differentially inhibits IFN-γ-inducible class II transactivator isoforms in both a TLR2-dependent and -independent manner. Immunol Cell Biol 90: 411-420.
-
Crossman DK (2007) Characterization of a novel acetylatransferase found only in pathogenic strains of Mycobacterium tuberculosis. Univ of Alabama: Tuscaloosa, AL.
-
Siddle KJ, Deschamps M, Tailleux L, Nédélec Y, Pothlichet J, et al. (2014) A genomic portrait of the genetic architecture and regulatory impact of microRNA expression in response to infection. Genome Res 24: 850-859.
-
Andraos C, Koorsen G, Knight JC, Bornman L (2011) Vitamin D receptor gene methylation is associated with ethnicity, tuberculosis, and TaqI polymorphism. Hum Immunol 72: 262-268.
-
Moores RC, Rand L, Elkington PTG, Friedland JS (2012) MMP-1 expression in tuberculosis is regulated by histone acetylation. in Department of Infectious Diseases & Immunity, Imperial College, London UK.





