GLI family zinc finger 3 (GLI3) has been shown to play a critical role in the regulation of Hedgehog signaling, with important implications in embryonic and postnatal development. However, the molecular characteristics of GLI3 gene and its expression profile in pig tissues have yet to be elucidated. In this study, the full-length cDNA sequence of the GLI3 gene from the lung of landrace pig was cloned and characterized using RT-PCR/RACE-PCR. The GLI3 full-length cDNA sequence was 6,064 bp, consisting of 4,761 bp of coding sequence encoding 1,586 amino acids with a molecular mass of 168.91 kDa. The 5′ UTR and 3′ UTR were 120 bp and 1,183 bp in length, respectively. Sequence alignment and phylogenetic analysis revealed a high similarity of the pig GLI3 amino acid sequence to that of other mammalian species. Furthermore, qRT-PCR analysis revealed that the GLI3 transcript was ubiquitously expressed in all tissues tested in this study, including the heart, liver, spleen, lung, brain, tongue and muscle. These results will provide a molecular basis for further studies of porcine GLI3 gene function.
GLI3, cDNA clone, Sequence analysis, Landrace pig
CDS: Coding Sequence; ORF: Open Reading Frame; pI: Isoelectric Point; qRT-PCR: quantitative Reverse Transcription Polymerase Chain Reaction; UTR: Untranslated Region; GLI3: GLI Family Zinc Finger 3; bp: Base Pair(s); cDNA: DNA Complementary to RNA; KDa: KiloDalton(s); RACE: Rapid Amplification of cDNA Ends
Hedgehog (Hh) signaling has been previously demonstrated to play a critical role in embryonic patterning, organ development and tumor growth [1-7]. Hh signaling pathway is well conserved from Drosophila to human. This pathway involves several essential components, including extracellular Hh ligands (Shh, Ihh or Dhh), membrane receptors Patched (Ptc) and Smoothened (Smo), and intracellular transcription factors, GLIs [8]. In vertebrates, the zinc finger-containing GLI proteins, including GLI1, GLI2, and GLI3, function as transcription factors that regulate the Hh signaling cascade. GLIs proteins have been shown to be invloved in embryonic patterning, organ development, and homeostasis [9].
Among these three transcription factors, GLI3 has become of great interest in the field due to its bifunctional nature. In the presence of Shh signaling, full-length GLI3 functions as a transcriptional activator (GLI3A). However, in the absence of Shh signaling, GLI3 is partially proteolyzed into a truncated transcriptional repressor (GLI3R) to block the expression of downstream target genes [10,11]. In addition, there is substantial evidence demonstrating that congenital mutations in the human GLI3 gene are associated with numerous clinical phenotypes [12-16].
Studies on genetically modified GLI3 mouse models have promoted insights into understanding of the molecular role of GLI3 in genetic disorders and development [17-20]. Especially, mutant mice lacking GLI2/GLI3 cause lung, oesophagus and trachea, indicating GLI3 plays a crucial role in foregut formation [19]. However, it is important to note that mouse model systems differ significantly from humans in many aspects, specifically physiological traits and gene expression. Thus, mouse model systems do not accurately represent conditions in humans and large animal models remain urgently needed [21,22]. Pig models possess potential advantages over rodents for biomedical research as their anatomy, physiology, and genetics are similar to humans [23,24]. Thus, GLI3 genetically modified pigs could serve as a better animal model to study the effect of GLI3 on embryonic and postnatal development. However, a complete genomic sequence harboring the porcine GLI3 gene sequence does not currently exist. In addition, the pig GLI3 transcript sequences are especially underrepresented in publicly available databases.
In the present study, we aim to obtain the full-length cDNA sequence of the GLI3 gene from landrace pig, analyze the structure of the deduced amino acid sequence, and investigate the expression of GLI3 gene in different pig tissues. This study will provide the molecular basis for further investigation of the landrace pig GLI3 gene function.
Fetal Chinese breed of landrace pigs were obtained from a pig breeding farm in Nanjing, China. Tissue samples were collected from the heart, liver, spleen, lung, kidney, muscle, and tongue by snap-freezing the tissue in liquid nitrogen, transported to the laboratory and stored at -80 ℃ until RNA extraction. All animal experiments were performed according to the guidelines approved by the Animal Ethics Committee of Nanjing Medical University.
Total RNA from different tissues, including the heart, liver, spleen, lung, brain, tongue and muscle from 110-day-sold fetal landrace pigs was isolated from approximately 100 mg of the tissue samples using Trizol reagent according to the manufacturer's instructions (Invitrogen, Carlsbad, USA) and purified using RNeasy columns (Qiagen, Hilden, Germany). For quantitative RT-PCR analysis, first strand cDNA was synthesized from purified total RNA using HiScript®II QRT SuperMix (Vazyme, Nanjing, China) following the manufacturer's protocol. Briefly, reverse transcription was performed in a 10 ΜL reaction volume containing 0.5 Μg of total RNA treated by 4 × gDNA wiper Mix at 42 ℃ for 2 min, 2.0 ΜL of 5 × HiScript® II qRTSuperMix II. Thermal cycling was carried out with incubation at 25 ℃ for 10 min, followed by 30 min incubation at 50 ℃, and a final step at 85 ℃ for 5 min. The synthesized first strand cDNA was stored at -20 ℃ until use.
To confirm the middle sequence of GLI3 mRNA, overlapping forward and reverse primer pairs (listed in Table 1) were designed with Primer 3 software [25]. RT-PCR reactions were carried out in a 25 ΜL reaction volume using ExTaq DNA polymerase (Takara, Dalian, China) with a pre-denaturation step at 94 ℃ for 5 min, followed by 35 cycles of 94 ℃ for 30 s; 60 ℃ for 30 s; 72 ℃ for 3 min, and a final step of 10 min at 72 ℃. To obtain the full-length porcine GLI3 cDNA sequence, the 5′ and 3′- ends of theGLI3cDNA sequence were amplified using a SMARTer® RACE 5′/3′ Kit (Clontech, Mountain View, USA) following the manufacturer's instructions. PCR amplification was carried out using gene-specific 5′ or 3′ primers (Table 1) paired with the Universal Primer Mix (UPM) under the following condition: 25 cycles of 94 ℃ for 30 s, 68 ℃ for 30 s, and 72 ℃ for 3 min. All the resultant PCR products (including 5′ RACE, 3′ RACE and the middle fragment PCR products) were examined by 1% agarose gel electrophoresis, and recovered using an Agarose Gel DNA Purification Kit (TIANGEN, Beijing, China). The products were cloned into the pMD-18T vector (TaKaRa, Dalian, China) and ten to fifteen amplicons were sequenced to ensure base accuracy by Genscript Co., Ltd. (Nanjing, China).
Table 1: Oligonucleotide primers used for GLI3 gene cloning. View Table 1
The full-length GLI3 mRNA sequence was assembled using Snap gene software (GSL Biotech LLC, Chicago, USA). Open reading frame of GLI3 was predicted using the NCBI/ORF Finder computational tool (https://www.ncbi.nlm.nih.gov/orffinder/). The theoretical molecular weight and pI of GLI3were calculated using Compute pI/Mw (http://web.expasy.org/compute_pi/). Signal peptide sequences were predicted using the Signal P 3.0 server (http://www.cbs.dtu.dk/services/SignalP/). SOPMA (http://npsa-pbil.ibcp.fr/) was used to predict the secondary structure of GLI3. The PSORT II web-based program (http://psort.hgc.jp/form2.html) was used to predict the subcellular localization of the GLI3 protein. The O-glycosylation sites, N-glycosylation sites and phosphorylation sites of the GLI3 protein were predicted using NetOGlyc 3.1 (http://www.cbs.dtu.dk/services/NetOGlyc/), NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/) and NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos-2.0/), respectively. The functional annotation of potentially conserved motifs was analyzed using SMART databases (http://smart.embl-heidelberg.de).
The accession numbers referring to GLI3 sequences from different animals in GenBank are listed in Table 2. A multiple sequence alignment of the GLI3 amino acid sequences was carried out using Clustalw2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The unrooted phylogenetic tree constructed from the alignment was generated based on neighbor-joining method with 1000 bootstrap replicates using Molecular Evolutionary Genetic Analysis (MEGA) software version 7.0 (http://www.megasoftware.net/).
Table 2: List of GLI3 gene sequences analyzed in this study. View Table 2
The expression level of GLIs mRNA in different tissues was assessed using quantitative Reverse Transcription PCR (qRT-PCR). The primers used for qRT-PCR were designed using Primer 3 software and synthesized by Genscript (Nanjing, China). The primers used for these experiments are listed in Table 3. PCR reactions were carried out in a 10 ΜL volume containing 1 ΜL of diluted cDNA template, 5 ΜL of Fast Start Universal SYBR Green Master (Roche, Penzberg, Germany), 0.66 ΜL of each forward primer and reverse primer. The qRT-PCR reactions were carried out on a LightCycler® 96 (Roche, Penzberg, Germany) apparatus with 1 cycle at 95 ℃ for 10 min, followed by 40 cycles at 94 ℃ for 10 s, 60 ℃ for 30 s. PCR amplification specificity was evaluated by the melting curve analysis. All samples were amplified in triplicates and ACTB was used as an endogenous reference gene. The relative gene expression levels of GLIs were then calculated using the 2−ΔΔCt method [26].
Table 3: Primer used for qRT-PCR. View Table 3
The predicted porcine GLI3 gene (GenBank: XM_013985838.1) and 15 GLI3 sequences from different animal species were collected when the gene symbol GLI3 was queried against the GenBank database (Table 2). Comparison of the 15 GLI3 sequences from different species revealed a striking degree of conservation in the length of exons 3-14 (Table 2). In contrast, the predicted porcine GLI3 gene sequence corresponding to the human GLI3 exons 10-13 was different in length from that of the other 15 GLI3 genes. Sequence analysis of the predicted porcine GLI3 protein identified the presence of a putative single zinc finger motif belonging to the C2H2 subclass. However, the GLI family is characterized by the presence of five tandem zinc fingers. Taken together, we inferred that the predicted porcine GLI3 gene was incomplete and invalid, partly due to gaps within the porcine genomic sequences that harbor the GLI3 locus. Thus, we utilized RT-PCR and RACE techniques in this study to determine the full-length mRNA sequence of the porcine GLI3 gene using cDNA generated from the lung of a landrace pig as a template, for the GLI3 gene expression is relatively high in lung than that in others organs or tissues in porcine.
An 899 bp fragment spanning from exon 9 to 13 was obtained by RT-PCR using primers GLI3-F1 and GLI3-R1. Based on the sequence of the 899 bp segment and the predicted porcine GLI3 gene sequence, the middle portion of the GLI3 mRNA sequence was determined using six overlapping primer pairs (Table 1). Subsequently, an 839 bp fragment and a 1,352 bp fragment were obtained using 5′ RACE PCR and 3′ RACE PCR, respectively. As a result, a 6,064 bp pig GLI3 mRNA sequence was obtained by aligning all of the above-mentioned fragments. This sequence consisted of 120 bp 5′ UTR, a 1,183 bp 3′ UTR and an ORF 4,761 bp in length (Figure 1A and Figure 1B). Our analysis showed that all of the exons of porcine GLI3 are the same sizes as those of GLI3 from 15 other species, with the exception of exons 1 and 15 (Table 2). This indicates that GLI3 has been well conserved throughout vertebrate evolution. The pig GLI3 cDNA sequence cloned in this study has been submitted to NCBI and has been assigned a GenBank accession number of KX768758.
 Figure 1: Nucleotide and amino acid sequence of GLI3 cloned from landrace pig. The nucleotide sequence is represented in black font and the amino acid sequence is represented in blue font. View Figure 1
Figure 1: Nucleotide and amino acid sequence of GLI3 cloned from landrace pig. The nucleotide sequence is represented in black font and the amino acid sequence is represented in blue font. View Figure 1
The deduced GLI3 was predicted to have a molecular mass of 168.91 kDa and an Isoelectric point (pI) of 7.2. A subcellular distribution analysis of the GLI3 protein by PSORT II predicted that 82.6% of the total protein may exist in the nucleus, 8.7% may be in the cytoplasm, 4.3% could be in vesicles of the secretory system, and 4.3% could be in the endoplasmic reticulum. Neither an N-terminal signal peptide nor a trans-membrane region was detected using the PSORT II software. Six N-glycosylation sites (Asn598, Asn722, Asn829, Asn888, Asn1048, Asn1072) and 159 phosphorylation sites, including 121 Ser residues, 24 Thr residues, and 14 Tyr residues were predicted to be present in GLI3 protein (Figure 2A and Figure 2B). The secondary structure of the GLI3 protein was predicted to be predominantly random coil (72.51%), followed by α-helix (14.56%), and β-fold (12.93%) (Figure 2C). Prediction of protein domains by the SMART program (http://smart.embl-heidelberg.de/) revealed that the porcine GLI3 protein was comprised of five tandem zinc finger motifs at positions 480-505, 513-540, 546-570, 576-601 and 607-632, similar to that observed in human GLI3 protein.
 Figure 2: Amino acid sequence characterization and phylogenetic analysis of GLI3 (A) GLI3 protein with six predicted N-glycosylation sites; (B) GLI3 protein with 159 predicted phosphorylation sites; (C) The predicted secondary structure of the GLI3 amino acid sequence. The blue lines represent α-helix, the red lines represent extended strand, the green lines represent β-turn, and the purple lines represent random coil; (D) Phylogenetic analysis of the amino acid sequences of GLI3 from pig and other species. The tree was constructed using the MEGA 7 software by Neighbor Joining method with 1000 bootstrap replicates. View Figure 2
Figure 2: Amino acid sequence characterization and phylogenetic analysis of GLI3 (A) GLI3 protein with six predicted N-glycosylation sites; (B) GLI3 protein with 159 predicted phosphorylation sites; (C) The predicted secondary structure of the GLI3 amino acid sequence. The blue lines represent α-helix, the red lines represent extended strand, the green lines represent β-turn, and the purple lines represent random coil; (D) Phylogenetic analysis of the amino acid sequences of GLI3 from pig and other species. The tree was constructed using the MEGA 7 software by Neighbor Joining method with 1000 bootstrap replicates. View Figure 2
The alignment of the coding region of 16 GLI3 gene sequences by CLUSTALW2 revealed a high identity amongst the detected species, with a minimum value at 80%. In addition, the deduced pig GLI3 protein shared 90.75%, 86.45%, 86.7%, 86.77%, 87.49%, 89.17%, 87.36%, 85.03%, 83.3%, 85.48%, 89.45%, 88.6%, 90.69%, 91.08%, and 85.29% amino acid sequence similarity to the GLI3 protein of Bos Taurus, Chlorocebus sabaeus, Homo sapiens, Pan troglodytes, Canis lupus familiaris, Felis catus, Oryctolagus cuniculus, Cavia porcellus, Mus musculus, Loxodonta africana, Equus caballus, Odobenus rosmarus, Bubalus bubalis, Lipotes vexillifer, and Trichechus manatus, respectively.
To clarify the evolutionary relationship between porcine GLI3 and other animal GLI3 molecules, a phylogenetic tree was constructed using MEGA7 software with the Neighbor Joining (NJ) method (Figure 2D). These results indicated that landrace pig GLI3 clustered with its homologs from 15 other species. Specifically, it revealed that porcine GLI3 possessed the highest homology with that from Bos Taurus and the lowest homology with that from Papio anubis.
The gene expression profile of GLI3 in different tissues, including the heart, liver, spleen, lung, brain, tongue and muscle from 110-day old fetal landrace pigs was monitored using qRT-PCR (Figure 3). The results showed that GLI3 was constitutively expressed in all tissues examined. However, expression levels were different depending on the tissue. High GLI3 gene expression levels were detected in the kidney and brain, while low expression levels were detected in the muscle and liver. In order to differentiate the accumulation in transcript levels of GLI3 with that of the other two GLI family members, the expression pattern of GLI1 and GLI2 were also investigated in the same tissue samples. GLI1 and GLI2 mRNA were also detected in all tissues tested. However, GLI1 mRNA expression was found to be significantly higher in the kidney compared to other tissues (p < 0.05). Overall, compared to GLI1 and GLI2, GLI3 showed higher expression levels in all tissues, with the exception of the liver where the GLI1 transcript was found to be expressed slightly higher.
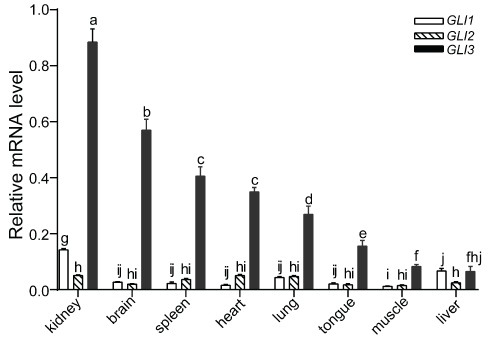 Figure 3: Relative mRNA expression levels of GLIs in various tissues from landrace pig. The expression levels were normalized to ACTB using 2−ΔΔCt method [27]. Values are mean ± SD. Significant differences in gene expression were determined by ANOVA. Means values indicated by the same letter were not significantly different (p < 0.05). View Figure 3
Figure 3: Relative mRNA expression levels of GLIs in various tissues from landrace pig. The expression levels were normalized to ACTB using 2−ΔΔCt method [27]. Values are mean ± SD. Significant differences in gene expression were determined by ANOVA. Means values indicated by the same letter were not significantly different (p < 0.05). View Figure 3
GLIs include a total of three family members. GLIs proteins have been shown to mediate the Sonic hedgehog (Shh) signaling pathway and play critical roles in embryonic patterning, organ development, and homeostasis [9]. Moreover, lung development was completely blocked in mutant mice lacking both GLI2 and GLI3 function [19]. As pigs have become ideal large animal models in biomedical studies, it is promising to harvest humanized lung organs from GLI2/GLI3 knockout pigs via interspecies blastocyst complementation [27]. To date, only GLI1 and GLI2 of pig have been identified [28,29]. In this study, we reported the cloning of landrace pig GLI3 for the first time.
The cloned full-length sequence of the GLI3 gene is 6,064 bp in length. This gene contains 4,761 bp of Coding Sequence (CDS) which encodes a 1,586 amino acid protein, a 120 bp 5′ UTR and a 1,183 bp 3′ UTR. BLAST results demonstrated that the landrace pig GLI3 sequence exhibits high identity to the GLI3 counterparts of other species. Consistent with sequence information from human and mouse GLI3 [30,31], porcine GLI3 contains an in-frame UGA stop codon located 12 bp upstream of the AUG initiation codon. In addition, porcine GLI3 was also found to encode five zinc finger motifs, providing further evidence for the evolutionary homology of GLI3 across species. The pig GLI3 protein was determined to have a molecular mass of 168.9 kDa and a theoretical pI of 7.2. This protein was predicted to not possess an N-terminal signal peptide or a transmembrane domain, which is similar to human and mouse GL3 proteins [32,33]. Previous studies have demonstrated that GLI3 proteins are regulated by posttranslational modifications, including phosphorylation, ubiquitination, and methylation [34-38]. Notably, we identified a large number of predicted phosphorylation sites within the pig GLI3 protein using NETPhos analysis. This result suggests that phosphorylation could play a key role in the regulation of the trans-activity of pig GLI3 protein. Further studies are needed to elucidate key phosphorylation sites involved in GLI3 regulation in pigs.
Phylogenetic tree analysis revealed that the amino acid sequence of landrace pig GLI3 shared high sequence similarity to GLI3 orthologs from other mammalian species. These sequences were found to possess greater than 83% sequence identity for all of the species analyzed. These results indicated that the amino acid sequence of pig GLI3 was highly evolutionarily conserved. Thus, these findings may substantiate the use of the pig as an ideal model for large animal systems to study GLI3 gene function.
qRT-PCR results demonstrated that the GLI3 transcript was expressed ubiquitously in all fetal pig tissues examined in this study, including the heart, liver, spleen, lung, brain, tongue and muscle. This result suggested that theGLI3 gene, which encodes a putative zinc finger transcription factor, could play a vital role in early organ development in pigs. Compared with GLI1 and GLI2, GLI3 exhibited higher expression levels in all tissues studied, with the exception of the liver, emphasizing the developmental importance of GLI3 in pigs. In addition, among all of the tissue types tested, GLI3 expression was found to be the highest in the kidney and brain. However, in humans, while GLI3 was found to be expressed in various tissues, no expression was observed in the kidney or brain, in contrast to what was observed in pig [30]. These inconsistencies between human and pig in regard to GLI3 expression patterns in the kidney and brain could be due to differences between the species or the developmental stage at which tissues were sampled.
In conclusion, we successfully cloned full-length GLI3 cDNA sequence from landrace pig lung in this study. This cDNA sequence is 6,064 bp long and encodes a 1,586 amino acid protein. Several bioinformatics and phylogenetic tools were used to analyze both the nucleotide and amino acid sequence of the molecule and for the characterization of the gene. In addition, we also evaluated relative mRNA expression levels of GLI3 in various landrace pig tissues and organs. Our findings provided an important basis to enable future studies regarding the function and regulatory mechanisms of porcine GLI3.
This work was supported by a grant from the Jiangsu Key Laboratory of Xenotransplantation (BM2012116).