The present study was conducted to assess Single Nucleotide Polymorphism (SNP) of adiponectin gene (ADIPOQ rs2241766) variant SNP + 45 T > G with reference to thyroid status in the three study groups, namely non-obese, overweight and obese type 2 diabetics.
150 Type 2 diabetics of both genders who had visited the tertiary health care clinics during the stipulated study period were included. Based on BMI (18.50 to 24.99, 25 to 29.99 and ≥ 30 respectively), they were segregated into non-obese, overweight and obese (n = 28, n = 76 and n = 46 respectively). The cardio-metabolic risk factors were assessed through parameters including anthropometric and routine biochemical measures, besides glycemic status and insulin resistance. The divalent cations (Zn2+ and Mg2+) were quantitated in serum. Serum free T4, T3 and TSH were estimated to evaluate thyroid status. PCR- RFLP was carried out to decipher the gene polymorphism of ADIPOQ (rs2241766) T > G of exon 2 in Adiponectin gene with particular reference to the implications in personalised medicine.
A positive association was found between SNP + 45 T > G in exon 2 and T2DM in the study population. Data on genotyping among non-obese vs. overweight depicts association with LDL in TT (Homozygous) p = 0.0491, while in TG (Heterozygous) phenotype, a strong association was perceived with Insulin resistance (p = 0.006). Associations concerning TSH in TT p = 0.0339; Mg, p = 0.0145 and p = 0.0138 in TT & TG respectively in obese vs. overweight and T4 in TT with p = 0.0491 were observed in non-obese vs. obese T2DM.
SNP + 45 T > G gene polymorphism in adiponectin gene may be a cardinal feature in insulin resistance observed in type 2 diabetes mellitus for consideration, based on BMI. Also, the wild type (TT) depicted good association with TSH, T4 and LDL in obese vs. overweight, non-obese vs. obese and non-obese vs. overweight type 2 diabetics respectively to stimulate interest in personalised medicine.
Type 2 Diabetes mellitus, Insulin resistance, Triiodothyronine, Thyroxine adiponectin, Polymorphism, Cardio-metabolic factors
Diabetes mellitus is a well-recognised multifactorial metabolic disorder which has the virtual ability to affect organ systems in the body [1]. Citing the eighth edition of Diabetes Atlas 2017 published by the International Diabetes Federation (IDF), the number of diabetics in India is 74 million which would swell to 75 million by the year 2020, unless aggressive measures to curb this menace are effected in a sustained manner [2].
Type 2 Diabetes mellitus (T2DM) possesses nexus with genetic and extra genetic components characterized by insulin resistance and gross pancreatic ß cell dysfunction [3]. The variations in the genetic factor, termed Single Nucleotide Polymorphisms (SNPs) influencing the phenotypes need to be still effectively and comprehensively addressed in Asian-Indian populations. There are several instances to cite the association of adiponectin gene variants with T2DM and obesity. Alternately, ambiguity shrouds the definitive role of genetic polymorphism in adiponectin gene, as related to insulin resistance and more so in obese and non-obese study populations [4].
Studies on gene polymorphisms in adiponectin, as related to insulin resistance are available [5]. However, planned and comprehensive evidence-based studies on adiponectin polymorphism, in general and insulin resistance in relation to thyroid status in particular from South India are sparse. There is an urgent need to explore in detail, based on laboratory evidences, especially for South-Indian population, as linked to thyroid status in T2DM, since altered thyroid status is pronounced in T2DM [6]. Nineteen common polymorphisms of fourteen known candidate genes have thus far been analysed worldwide for their contribution to prevalence and incidence of glucose intolerance. This is portrayed in an epidemiological study on the Insulin Resistance syndrome [7]. ADIPOQ is one such 17 Kb gene whose gene is located on chromosome 3q27, a region that has already been implicated in susceptibility to type 2 diabetes and obesity. The gene comprises of three exons and two introns [8]. Adiponectin (MW 30 kDa) is secreted from the adipose tissue, chiefly the white adipose tissue and essentially comprises of 244 amino acid residues [9]. Adiponectin possess a wide gamut of anti-diabetic, anti-inflammatory and anti-atherogenic properties, as evidenced from the literature [10,11]. SNPs have been detected in ADIPOQ gene, which are associated with clinical disorders [12].
An Indian study depicted positive association of a particular polymorphism of the adiponectin gene, namely SNP + 45 T > G (rs2241766) with type 2 diabetes mellitus [13]. Although, altered thyroid profile is widely implicated in T2DM, it must be said in all fairness that limited documentation is available implicating adiponectin gene polymorphism in insulin resistance associated with altered thyroid status. Hence, in the light of the above considerations and based on the existing documentation, we had embarked upon the study to analyse the association of SNP + 45 gene polymorphism on Thyroid status in Type 2 Diabetes, as exemplified in the three study groups, namely non-obese, overweight and obese. This would possibly open vistas to apply pharmacogenomics/personalised medicine in insulin resistant type 2 diabetics, with special reference to thyroid status.
We took up the study based on an earlier paper published from India by Matharoo, et al. who had elicited the association of the genetic polymorphism, namely SNP + 45 (ADIPOQ rs2241766) of adiponectin, in the light of insulin resistance [14] and as observed in T2DM. Interestingly, the results of the study by Matharoo, et al. depicted an association that was observed between adiponectin SNP + 45 and increased risk of T2DM. Furthermore, the same study delineated the observation that a statistically significant association was observed between the anthropometric indices BMI and WC and T2DM. We primarily focused on gene polymorphism pertaining to SNP + 45 of adiponectin as linked to insulin resistance in the three groups of type 2 diabetics, namely non-obese, overweight and obese, based on a previous report from South India on SNP + 45 of Adiponectin (13), but we carried out the study with reference to thyroid status.
We also felt that since adiponectin levels and altered Thyroid status have been implicated in insulin resistance, there is an immediate need to link these two in the light of gene polymorphism of Adiponectin, but with reference to obese, non-obese and overweight type 2 diabetics. Studies on Asians have also delineated the link between SNP + 45 and metabolic syndrome [15]. The culmination of these would enable us to arrive at personalized medicine based on an objective pharmacogenomics approach to address thyroid status in insulin resistance associated with adiponectin, but as a function of body mass index.
Furthermore, controversy does exist with respect to the definitive role of adiponectin in thyroid status, though thyroid hormones and adiponectin are believed to be associated with each other independently of weight status. It was reported that adiponectin levels were higher in hyperthyroidism compared to hypothyroidism or euthyroidism, respectively [16,17]. Conversely Santini, et al. and Iglesias, et al. have reported that adiponectin levels were not significantly different in hyperthyroidism in comparison to control groups [18,19].
However, Altinova, et al. could not demonstrate a significant association between thyroid status and adiponectin levels [20]. Whereas, another study suggested that thyroid hormone levels are associated with adiponectin levels in healthy subjects [21].
In view of the prevailing scenario with reference to the role of adiponectin in thyroid status of type 2 diabetics and also due to the limited availability of the studies on gene polymorphism of adiponectin as related to insulin resistance in obese, non-obese and overweight type 2 diabetics, we proceeded to determine thyroid status in the present study.
This study was performed at a tertiary care hospital at Pondicherry, South India. The study included 150 type 2 diabetic subjects of both genders in the age group of 35-70 years who had attended the outpatient clinics. The research study was duly approved by the Research Advisory committee (RAC) and Institutional Human Ethics Committee (project No:Ph.D./2015/02 dt.5th June, 2015, signed by the Secretary IHEC). The study was conducted between January 2017 and July 2017.
The sample size n = 150 was computed by using CaTS (Centre for statistical genetics) power calculator with significance level 0.05% with a power value of 81%. The purpose of the study was explained in detail to the participants in the vernacular language and due consent obtained. The cases were included only on the absolute basis of clinically and biochemically confirmed type 2 diabetes mellitus. The diagnosis of T2DM on the study subjects was confirmed by duly qualified clinicians manning the outpatient clinics. The clinicians were in possession of recognised postgraduate qualifications and experience as laid down by the regulatory agency, in internal medicine and having proven experience in endocrinology. All patients were interviewed for eliciting comprehensive medical history, personal history of smoking/alcohol/substance abuse, and family history of diabetes mellitus and thyroid disorders.
Grouping/segregation was mandatorily based on BMI, into non-obese, overweight and obese which was enabled only after taking all of the study subjects: -confirmed type 2 diabetics (n = 150) into due consideration.
Care was taken to see that the patients were not billed for undergoing additional and special biochemical investigations other than those that were absolutely considered essential for rational management of T2DM, as per the institute policy.
Subjects with known thyroid and other endocrine disorders as well as those who had exhibited clinically significant neurological, cardiovascular, respiratory and gastrointestinal or any other majowm systemic ailments, malignancies were excluded from the study.
Two ml of venous blood samples were drawn into vacutainers containing EDTA for genetic polymorphism studies. Samples were stored at -70 ℃ until further analysis of DNA for monitoring single nucleotide polymorphism of adiponectin (ADIPOQ rs2241766 T > G of exon 2 in adiponectin gene). An additional three ml of venous blood was collected for estimating various biochemical parameters.
Fasting blood glucose was estimated by glucose oxidase-peroxidase method (GOD/POD); Fasting insulin was quantitated by automated chemiluminescence. Glycated haemoglobin was quantitated by HPLC method. Insulin resistance was measured based on the formula [22] HOMA - IR (Fasting plasma glucose (mmol/l) × plasma fasting insulin (m IU/l)/22.5).
Triacylglycerols in serum was measured by glycerol kinase method; Total cholesterol by enzymatic method; HDL cholesterol was enabled by polyanion precipitation. LDL cholesterol was computed by Friedwald equation, LDL cholesterol = Total cholesterol-(HDL cholesterol + VLDL) where VLDL = TAG/5; LDL particle size was quantitated using the surrogate marker (TAG/HDL), to denote small dense LDL. Zinc was estimated by colorimetric method based on the procedure described by Tetsuo Makino [23] and quantitation of magnesium was enabled by spectrophotometric assay using xylidyl blue. T3, T4 & TSH were quantitated based on automated electro chemiluminescence method.
High HOMA-IR was defined as HOMA-IR ≥ 2.69 [1,25].
Glucose (Fasting plasma, venous): 70-110 mg/dL; Insulin (Fasting plasma, venous): 0.7-9 µu/mL; Glycated Hemoglobin (HbA1c): goal is to keep the levels below 7%; Total cholesterol: 150-200 mg/dl; Triacylglycerols: 75-150 mg/dl; High Density Lipoprotein cholesterol: 30-60 mg/d; Free Triiodothyronine (FT3): 2-4.4 pg/ml; Free Thyroxine (FT4 level): 0.93-1.7 ng/ml; TSH: 0.27-4.2 µu/mL; Magnesium: 1.8-3 mg/dl; Zinc: 60-120 µg/dl.
Whole blood was used to isolate DNA and to facilitate the determination of genotype frequency of Adiponectin SNP + 45: Exon 2 of the ADIPOQ was suitably amplified by Polymerase Chain Reaction (PCR) and the genomic DNA was used for amplification.
The protocol for PCR was followed by Restriction Fragment Length Polymorphism (RFLP) denoting the three genotypes, namely wild type (TT), heterozygous type (TG) and homozygous variant type (GG) and these were based on a previous method, as applied on a south Indian population [12]. We had employed 2.5% agarose gel under optimal conditions for separation of the restriction digest. To enhance objectivity and uphold reproducibility and reliability of data in each batch, a few positive control samples for homozygous and heterozygous genotypes were also established.
Stringent quality control in the assessment of biochemical parameters was adhered to and internal quality control for the same was promulgated, based on the Quality Control (QC) samples provided by M/s Biorad, USA. External quality Assessment of routine biochemical parameters was enabled through the Clinical Biochemistry laboratory, Christian Medical College (CMC), Vellore, under the External Quality Assessment Scheme (EQAS).
As regards the distribution of the alleles of SNP + 45 of Adiponectin gene, it was tested for Hardy-Weinberg equilibrium (p < 0.05). Proportions of genotypes of alleles were compared through Pearson χ2 analysis, odds ratios (ORs) and 95% confidence intervals (CI).
The results of the sub group analysis (shown in Table 1) depict the gene polymorphism, as exhibited in the genotyping found between the two groups, viz. non-obese and overweight. The heterozygous pattern, namely TG (T > G) clearly depicted statistically significant values among Fasting blood sugar, Postprandial sugar, Glycated haemoglobin, Insulin and HOMA-IR, whereas in TT (homozygous), LDL and Mg emerged statistically significant.
Table 1: Genotyping among 2 groups (Non-obese vs. Overweight). View Table 1
In Table 2, the gene polymorphism as exhibited in the genotyping found among two groups, namely overweight and obese is shown. The heterozygous pattern, namely TG (T > G) clearly depicts statistically significant value for magnesium (p value 0.0138), (p value = 0.0145) in TT and TSH (p value = 0.0339) in TT.
Table 2: Genotyping among 2 groups (Obese vs. Overweight). View Table 2
Table 3 summarises the results of the SNP studied, namely SNP + 45, as compared between non-obese and obese. The values denote statistically significant association between thyroxine levels (p value = 0.0491) and the wild type.
Table 3: Genotyping among 2 groups (Non-obese vs. Obese). View Table 3
Our results indicate variation in gene polymorphism with reference to three subgroups, namely non-obese, overweight and obese as observed in type 2 diabetes mellitus and with specific nexus with IR and thyroid status. The magnitude and percentage of wild type and variants were computed among non-obese, overweight and obese T2DM patients as depicted in Table 4, Table 4a, Table 4b, Figure 1 and Figure 2. We calculated the genotype frequencies of SNP 45 T G of adiponectin gene in T2DM. Pearson Chi-square test was enforced in order to determine the strength of association between the variant form and incidence of type 2 diabetes. The variant form TG is found to be significantly associated with type 2 diabetes mellitus (P 0.01). Our study demonstrates that subjects in possession of the variant type show a greater risk (Odds ratio = 3.077) of having type 2 diabetes than those carrying the wild type in Obese vs. Overweight.
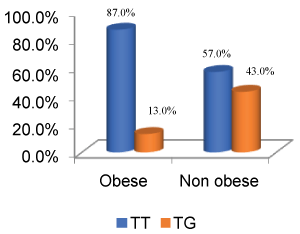 Figure 1: Depiction of % distribution of genetic polymorphism in ADIPOQ SNP + 45 in Obese vs. Non-obese (p value = 0.004, 95% CI = 1.601-15.613, odds ratio = 5).
View Figure 1
Figure 1: Depiction of % distribution of genetic polymorphism in ADIPOQ SNP + 45 in Obese vs. Non-obese (p value = 0.004, 95% CI = 1.601-15.613, odds ratio = 5).
View Figure 1
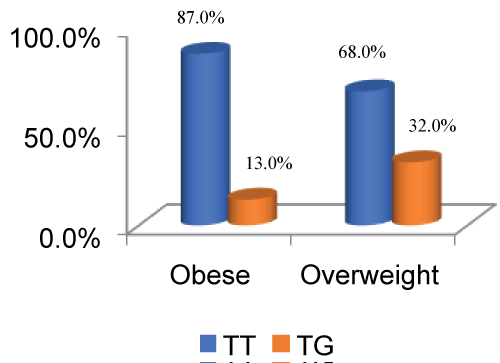 Figure 2: Depiction of % distribution of genetic polymorphism in ADIPOQ SNP + 45 in Obese vs. Overweight (p value = 0.021, 95% CI = 1.149-8.239, odds ratio = 3.077).
View Figure 2
Figure 2: Depiction of % distribution of genetic polymorphism in ADIPOQ SNP + 45 in Obese vs. Overweight (p value = 0.021, 95% CI = 1.149-8.239, odds ratio = 3.077).
View Figure 2
Table 4: Distribution of polymorphism-Overweight vs. Non-obese. View Table 4
Table 4a: Distribution of polymorphism-Obese vs. Non-obese. View Table 4a
Table 4b: Distribution of polymorphism-Obese vs. Overweight. View Table 4b
Table 4, Table 4a and Table 4b depict gene polymorphism as compared between the groups.
The distribution of wild type and heterozygous among obese and non-obese (P value = 0.004, 95% CI = 1.601-15.613, odds ratio = 5) is pictorially represented below in Figure 1, whereas Figure 2 indicates depiction of % distribution of genetic polymorphism in ADIPOQ SNP + 45 in Obese vs. overweight (p value = 0.021, 95% CI = 1.149-8.239, odds ratio = 3.077).
Figure 3 shows the agarose gel electrophoresis pattern obtained as a result of RFLP on amplified DNA enabled by PCR. The purified products emanating from PCR were subjected to restriction digestion utilising the restriction endonuclease, namely SmaI that culminated in the following fragment sizing patterns, as perceived through agarose gel electrophoresis (Figure 3). Wild type, wherein exon 2 of adiponectin, depicted as 45TT: It is to be noted that no cleavage of the intact 470 bp segment by SmaI was observed. The heterozygous variant type is denoted by 45TG: The restriction enzyme SmaI incises at the sequence CCC; GGG to show three fragments, as exemplified by the analytical performance, based upon agarose gel electrophoresis (470 bp, 336 bp and 134 bp respectively). The homozygous variant type is referred to as 45GG and SmaI incision at CCC; GGG sequence generated two cleaved fragments, as portrayed in agarose gel electrophoresis (336 bp and 134 bp respectively). However, our results unequivocally proved that the homozygous genotype did not manifest in the present study.
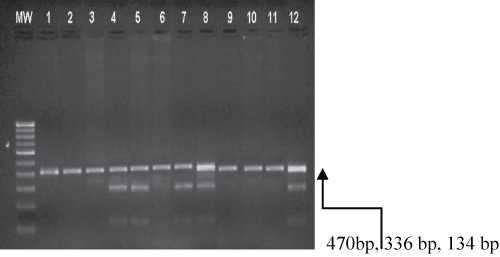 Figure 3: Agarose gel electrophoresis showing the PCR-RFLP pattern of adiponectin SNP + 45.
View Figure 3
Figure 3: Agarose gel electrophoresis showing the PCR-RFLP pattern of adiponectin SNP + 45.
View Figure 3
It is to be noted that the bands obtained in Figure 3 denote wild type TT (470 bp), heterozygous type TG (470 bp, 336 bp, 134 bp), MW - Molecular DNA marker (100 bp). Lanes 4, 5, 7, 8 & 12 - heterozygous TG (470, 336, 134 bp). Lanes 1, 2, 3, 6, 9, 10 &11 - Wild type TT (470 bp).
Adiponectin has been widely implicated in possessing anti-diabetic properties, besides other documented attributes that include anti-atherogenic and anti-inflammatory properties. Adiponectin gene (ADIPOQ) variability may affect the risk for type 2 diabetes mellitus (T2DM) and complications. An earlier report had implicated genetic variability on adiponectin gene as associated with extracellular levels of inflammatory and angiogenic markers [26]. However, these authors felt that further research is warranted to unequivocally elucidate the role of adiponectin in the development and/or progression of micro vascular disease in T2DM patients.
An elaborate understanding of adiponectin has been vividly described [27]. However, controversy still shrouds especially with reference to polymorphism of adiponectin gene in modulating IR. Two majowm polymorphisms of adiponectin gene, namely SNP + 45 and SNP + 276 have been documented, but with variegated actions governing Insulin sensitivity and IR [28]. It is well known that alterations in thyroid status have been observed, thanks to the global research findings on IR associated with T2DM. However, controversy still persists even in this area, since biochemical events synonymous with both hypothyroidism and hyperthyroidism have been reported by different groups of workers leading to a paradox. Furthermore, the available literature on thyroid status in insulin resistance does not take into consideration the holistic purview of including anthropometric, physiological, biochemical and molecular biology parameters, in the same study during a given timeline. In addition to the above-mentioned facts, very few studies from South India have implicated SNP + 45, in the light of insulin and thyroid status.
The gene polymorphism in Adiponectin, namely SNP + 45 T/G (rs2241766) has been considered as an important clinical feature to consider prior to pioglitazone treatment [29]. Our results also suggest that there is an excellent association between T > G and T2DM, especially when comparisons were made between non-obese vs. overweight and overweight vs. obese. Hence, gene polymorphism in Adiponectin T/G needs to be given greater impetus while planning pharmacologic modalities for overweight and obese diabetes. A meta- analysis of gene polymorphism in ADIPOQ gene +45 in the Asian population of type 2 diabetics augments further the cardinal role of SNP + 45 T/G [30]. It has been long known that thyroid abnormalities directly are linked to cardiovascular morbidity [31], as also to insulin resistance [32]. We were curious to study the effect of ADIPOQ gene polymorphism SNP + 45 T > G, in the light of these established findings. To our surprise, besides elucidating statistically significant association between SNP + 45 T > G and T2DM in non-obese vs. overweight and overweight vs. obese, there was a pronounced association even between the wild type TT and TSH in obese vs. overweight comparison. A strong association has been perceived with reference to T4 in wild type TT (obese vs. non-obese). These results point to relationship of Hypothalamus-Anterior Pituitary-Thyroid axis, based on BMI and gene polymorphism and we need to bear in mind, while planning personalised therapeutic strategies to combat cardiovascular morbidity, in addition to altered thyroid status in Insulin resistant type 2 diabetics.
Hsieh and Wang have evaluated circulating levels of adiponectin in patients with thyroid dysfunction prior to and following normalization of thyroid function with proper drug intervention. As per their study, the change in BMI was strongly correlated with changes in serum-adiponectin levels and changes in serum free thyroxine also correlated well with changes in BMI and serum adiponectin levels. In our present study, we had utilised BMI as the anthropometric measure which facilitated the advent of an additional group, namely overweight, in addition to obese and non-obese. This would not have occurred, had we included waist circumference (since we use waist circumference for segregating obese from non-obese) as an anthropometric measure. However, since we had used BMI to segregate the study subjects into obese, non-obese and overweight, we were able to compare specific groups for associations among markers of IR and gene polymorphism of adiponectin and tried to look for nexus with Thyroid status. Hsie and Wang further had performed multivariate-regression analysis that revealed BMI to be a statistically strong predictor for serum-adiponectin levels. However, the same study also revealed thyroid function level as another predictor. Although BMI is the most frequently used anthropometric measure, it must be said that waist circumference is used for assessing central obesity [33]. We strongly opine that when waist circumference is taken into consideration, the facet of overweight will not be revealed. However, when BMI is used as the anthropometric measure in the light of Insulin resistance, overweight category unfurls. But, at the same time we also feel that an inherent demerit of using BMI is that despite the fact that it may evolve the study groups into three, namely non-obese, overweight and obese, it may still miss out central adiposity (obesity), a forerunner to insulin resistance. This is because a person with normal BMI may still have central obesity. Hence, we suggest that for instance, if we need to compare IR, as related to thyroid comorbidity in borderline obesity, then it is imperative that we include BMI, not WC. This may also explain why there is difference in the interpretation of data with reference to the genotype TT (wild type) and TG (Heterozygous) by earlier groups of workers. A revelation that has evolved from our present study is that even the wild type TT is associated only in overweight which would go unreported, if WC is taken as the anthropometric measure to determine obesity. Also, several workers have used BMI as the indicator of obesity but have simply reported as < 30 or ≥ 30. This may not be the correct stand to take as the priority lies in restoring Thyroid status associated with IR as perceived in overweight group, since it would turn out to be much more prophylactic in addressing the overweight aggressively which otherwise would eventually exacerbate thyroid comorbidity in insulin resistance, synonymous with obesity. Hence, from our study, it is clear that BMI would be a better status indicator of obesity, in addition to its capacity that lends itself to the unfurling of the overweight group.
There is a high prevalence of hypothyroidism in patients with extreme obesity. High levels of TSH contribute to elevated pro-inflammatory and cardiovascular risk markers, thereby increasing the risk for development of cardiovascular diseases [34]. Our results corroborate with the findings of other workers on different ethnic groups [5,7,11,13,26-28,35]. In the STOP-NIDDM trial [35], the G-allele of SNP + 45 was associated with 1.8-fold risk for type 2 diabetes mellitus. The present study of ours correlates with the findings of the STOP-NIDDM trial. Our results suggest that the haplotypes containing G allele, i.e. TG is significantly associated with a 3.077-fold increase for type 2 diabetes. Nakatani, et al. [36] in 2005 showed association of two single-nucleotide polymorphisms (SNP + 45T G, SNP + 276G T) with type 2 diabetes in the Japanese population. In their study, SNP + 45 was associated with insulin resistance and obesity. SNP + 276, another gene polymorphism showed a stronger association with IR and BMI as evidenced in the same study [37].
However, we were curious to study the role of SNP + 45 more so in the light of IR and Thyroid status in obese, non-obese and overweight type 2 diabetics, as it has not been studied in detail hitherto. Our contention is that it may be worth administering thyroid analogues and receptor modulators that would not only alleviate IR but also correct underlying early thyroid pathology. Our study would also explain that polymorphism alone in adiponectin SNP + 45 would not govern insulin and thyroid status in type 2 diabetics because even the wild type gene in exon2 of adiponectin is associated with T4 and TSH in non-obese vs. obese and obese vs. overweight type 2 diabetics respectively. More detailed studies need to be undertaken in this regard.
Earlier studies had implicated serum magnesium in IR associated with type2 diabetes mellitus [38,39]. In our present study also magnesium is associated both with wild type and heterozygous SNP + 45 of adiponectin gene in obese vs. overweight. We believe that the additional information is missing here which could be even directly attributed to insulin receptor, since it is well known that the insulin receptor gets down regulated in obese type 2 diabetics. Hara, et al. in 2002 [11] cited evidence of an association between polymorphisms at positions +45 and +276 in the adiponectin gene and type 2 diabetes respectively. Their study depicted the fact that only GG haplotype is associated with insulin resistance. Neither homozygous nor heterozygous carriers of TG haplotype had an association with insulin sensitivity in the Japanese population [5]. However, a few studies have shown no association between SNP + 45 T > G of adiponectin gene and IR [40-42]. This may be attributed to variation, in the ethnic groups. Also, we feel that the use of anthropometric measures as well as the assessment of the hypothalamus-anterior pituitary-Thyroid axis may offer newer vistas in clinical chemistry.
It is believed that SNP + 45 T > G could trigger insulin resistance enroute to the development of type 2 diabetes mellitus, possibly through changes in mRNA stability, levels of adiponectin and eventually reduced plasma adiponectin concentrations. We opine that the presence of SNP + 45 T > G could be one of risk factors for developing type 2 diabetes mellitus, besides being associated with altered thyroid status.
Direct evidence to support SNP + 45-induced regulation of adiponectin expression is still lacking and warrants further investigation. In another study by Yu, hyperthyroidism was associated with a 95% increase in adiponectin. Adiponectin was negatively correlated with BMI, total cholesterol and plasma triacylglycerols. Surprisingly, adiponectin was positively correlated with insulin, thereby suggesting that thyroid disease may be accompanied by changes in adipokines, which may contribute to the phenotype expressed [43]. An earlier study on TG/GG genotype in obese and non-obese non-diabetics also revealed that SNP + 45 T > G genotype was associated with enhanced fasting blood glucose, Insulin levels and Insulin resistance with accompanying low levels of total adiponectin in obese [44]. Thirty-two obese Japanese women were treated by meal replacement with a low-calorie diet for 8 weeks and asked to maintain their habitual lifestyle. Following the treatment, the extent of decrease in waist circumference was greater in the subjects with the G/G or G/T genotype of SNP276 than in those with the T/T genotype (p = 0.026).
As for SNP45, the extent of decrease in triglyceride levels was greater in the subjects with the T/T genotype than in those with the T/G genotype [45]. It is well known that gene polymorphism could vary with the population under consideration. Whereas, the results of a meta-analysis suggested that SNP + 45 G allele might be a susceptibility allele for T2DM in the population, as elaborated by Li and Li [46]. In another meta-analysis report, the results indicated the susceptibility of obesity as a function of adiponectin gene polymorphism [47].
Our present study also clearly depicts that even the wild type SNP + 45 in exon 2 namely TT could itself be used as a molecular indicator of altered thyroid status in Insulin resistant type 2 diabetics, as studied in overweight and obese. Our study further denotes differential effects of gene polymorphism with reference to SNP + 45 in obese, non-obese and overweight type 2 diabetics. An earlier paper published from our laboratory had implicated TAG/HDL ratio (surrogate marker of small dense LDL) and thyroid hormone levels in IR as observed in T2DM. We had concluded from that study that small dense LDL could be used as a reliable marker for IR, with accompanying alterations in thyroid status in overweight type 2 diabetics [48]. Our present study indicates LDL to be statistically significant while comparing non-obese and overweight T2DM. Hence, we strongly advocate that both wild and heterozygous SNP + 45 should necessarily be studied alongside the anthropometric parameter, namely BMI in order to delineate the overweight group in addition to the other two groups namely non-obese and obese that is frequently reported in the literature. Furthermore, our study demonstrates association with Thyroid hormones.
We opine that it may be worthy to afford direct measurement of adiponectin expression in white adipose tissues obtained from individuals with the SNP + 45 genotype and its association with plasma fasting glucose level, insulin level, insulin resistance combined with the dynamic function tests on Hypothalamus-pituitary-thyroid axis that would provide greater and reliable insight into the association of SNP + 45 as far as the pathogenesis of thyroid comorbidity in type 2 diabetes mellitus is objectively concerned.
1. Small sample size.
2. Lack of homogeneity in the local population which is a mixture of urban, semi urban and rural.
3. Lack of comparison between SNP + 45 and +276 Adiponectin gene polymorphism with reference to thyroid co-morbidity associated with insulin resistance.
It is concluded that the presence of G allele of Adiponectin +45T/G is a significant predictor of Thyroid function, based on BMI and subsequent grouping into non-obese, obese and overweight type 2 diabetics. Based on BMI, it is possible to assess thyroid status in type 2 diabetics as a function of gene polymorphism of adiponectin SNP + 45. Even it is possible to assign role for wild type (TT) of adiponectin exon 2 as a function of thyroid status. Also, it can be speculated that the polymorphism +45T/G may act as a strong marker of thyroid comorbidity in the studied Pondicherry population (South Indian) which is a mixture of urban, semi-urban and rural. Thyroid comorbidity is quite common in T2DM and it is quite possible that the Hypothalamus-pituitary-thyroid axis may play a cardinal role in the differential association between Adiponectin gene polymorphism and T2DM in obese, overweight and non-obese diabetics.
The findings showed that the prevalence of metabolic abnormalities and their clustering into MS increased with overweight and that BMI is a significant predictor of MS in this age group. These findings suggest early screening for metabolic abnormalities in adult T2DM and for thyroid function and also of early weight management intervention strategies. Furthermore, the judicious use of anti-diabetic drugs such as pioglitazone that influence adiponectin concentration could also be evaluated to restore thyroid status when used in combination with other conventional modalities used in the treatment of thyroid diseases.
Thus, the results of this current study represent the association of SNP 45 T > G genotypes of the adiponectin gene with insulin resistance and associated thyroid status differentially in the three groups, namely non-obese, overweight and obese type 2 diabetics. It is recommended that a combination of antidiabetic drugs, hypolipidemic drugs and thyroid receptor modulators could be envisaged to restore euthyroid status, besides alleviating IR, a factor that would assume great relevance in pharmacogenomics and personalised medicine, especially related to obese type 2 diabetics.
The findings that have emerged from the study could provide an opportunity to understand better roles and impact of adiponectin gene polymorphism in insulin resistance associated with altered thyroid status and could herald improved and objective modalities to manage overweight or obesity in individuals with uncontrolled T2DM. Hence, these findings may provide novel insights to control glycaemia and address insulin resistance progression objectively that may help improve thyroid status too. Furthermore, this will help clinicians to screen overweight patients much earlier, thus preventing possible serious macrovascular consequences either because of progressive insulin resistance or thyroid co-morbidity or both. We suggest that the use of novel thyroid receptor ligands with reference to gene polymorphism of Adiponectin would usher in fresh approach based on personalised medicine.
The authors wish to express their appreciation to the study participants and thank Prof. N Ananthakrishnan, Dean (Research), Sri Balaji Vidyapeeth. The authors also sincerely thank Dr Vettriselvi, Associate Professor, Department of Human genetics, Sri Ramachandra Medical College and Research Institute. Porur, Chennai.
No funding from external agencies.
The authors declare that there is no conflict of interest in this study.
R. Jayanthi, the first author was comprehensively involved in bench work. A.R.Srinivasan, the second author as well as the corresponding author contributed to the study design and associated logistic work, whereas, G.Niranjan, the third author had given suggestions for discussion, besides helping with the statistics.