Human cutaneous warts are prevalent worldwide and are caused by Human Papillomavirus. Most cutaneous warts can be clinically diagnosed without any auxiliary examination, but additional tests are required to confirm the infection of Human Papillomavirus in some atypical cases. The aim of this study was to measure the applicability of HPV DNA detection in cutaneous warts in a clinical setting.
A total of 345 skin surface swab samples were collected from clinically diagnosed common wart (n = 166), plantar wart (n = 142), flat wart (n = 5), both common and plantar warts (n = 15) and common and flat warts (n = 1). DNA extraction and amplification were carried out using an established PCR, FAP primer pair-based method to detect HPV DNA. We measured HPV DNA negative samples by PCR with β-globin primer pair to confirm the availability of DNA.
This study included 338 efficient DNA samples. The prevalence of HPV in common wart was 95.1% (154/162), plantar wart was 89.2% (124/139), flat wart, and the combination of common and plantar wart, common and flat wart were all 100% (6/6, 15/15, 1/1). The overall positive HPV detection rate in cutaneous warts was 93.2%.
These results suggest that the employed method is sensitive for HPV detection, with a total detection rate of 93.2%, and therefore it can be used as auxiliary examination for cutaneous warts.
Cutaneous warts, Human Papillomavirus (HPV), Polymerase Chain Reaction (PCR), FAP primer
Human Papillomaviruses (HPVs) are a large family of small, non-enveloped, and double-stranded DNA viruses with more than 180 subtypes that exclusively infect epithelial cells and are the cause of benign epithelial proliferation.
HPV also has the capacity to stimulate cells to premalignant and malignant transformation [1]. HPVs are divided into mucosal and cutaneous types [2]. Mucosal types target the mucosal membranes which in turn causes cervical neoplasia in women and anogenital warts in adults and children. While cutaneous types infect the squamous epithelium of the skin and leads to a diverse range of epithelial lesions, including common warts, plantar warts, flat warts, etc. [3].
Cutaneous warts are routinely diagnosed according to the clinical manifestation and clinical features. In cases with diagnostic uncertainty, additional tests for confirmation of the diagnosis are required; as such effective methods can provide proof of HPV infection. Current methods, e.g. In Situ Hybridization (ISH), Immunohistochemistry (IHC), Type-Specific Multiplex Genotyping (TS-MPG), Luminex Xmap Technology and Polymerase Chain Reaction (PCR) have been exploited. However, both ISH and IHC are costly and time-consuming [4]. IHC using anti-HPV antibody is less sensitive. TS-MPG assay can only detect 25 beta-HPV subtypes which predominantly infect skin tissue [5]. Luminex xMap technology allows rapid, sensitive and specific nucleic acid detection for 33 HPV-associated subtypes, again is costly and not readily available in the clinical setting [3,6]. A PCR-based method was developed that different primers were specifically designed based on the relatively conserved regions of the L1 open reading frame of HPV. Usually, there are three primers for the detection of HPV DNA by PCR method, MY09/MY11 followed by GP5+/GP6+ for α-HPV, CP65/CP70 followed by CP66/CP69 for β-γ-HPV, and FAP59/FAP64 for α-β-γ-HPV [2]. The FAP primer set allowing detection of up to 65 HPV subtypes improved HPV detection rate compared with other PCR-based methods [7]. We tested the sensitivity and applicability of the above-mentioned PCR method on scrape samples from several clinically diagnosed HPV infected skin conditions, in the aim of employing this simple and cheap method in aiding the diagnosis of clinical uncertain HPV infections.
Skin surface swab samples from clinically diagnosed cutaneous warts (common wart, plantar wart, flat wart) were collected from the Department of Dermatology, First Hospital of China Medical University, Shenyang, China from February 2012 to 2014. The samples were stored at -20 °C until analyzed. An informed consent was obtained from all enrolled subjects. The patients' clinical information is shown in Table 1.
All the swab specimens underwent DNA extraction using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the protocol described by the company. The presence of amplifiable DNA was verified using PCR with primers specific for β-globin gene.
25 μl final volume of the PCR reaction mixture contained 1.0 mM of each primer, FAP 59 (5' TAACWGTIGGICAYCCWTATT 3') and FAP 64 (5' CCWATATCWVHCATITCICCATC 3') [7], 12.5 μl GoTaq Colorless Master Mix (Promega, Mannheim Germany), 4.5 μl of nuclease-free water and 6 μl of extracted DNA sample. The cycling condition for PCR were denaturing at 94 °C for 1.5 min, annealing at 50 °C for 1.5 min, extension at 72 °C for 1.5 min for a total of 45 amplification cycles. The first cycle was preceded by 10 min denaturation at 94 °C. The last cycle was followed by an additional 7 min extension at 72 °C. PCR products were identified by electrophoresis in a 2% agarose gel containing TBE buffer and ethidium bromide. An amplicon yield of 480 bp DNA band used by FAP primer pair is considered as positive HPV DNA. An amplicon yield of 268 bp DNA band produced by β-globin primer pair was considered positive for efficient human DNA.
A total of 345 skin swabs from 329 patients clinically diagnosed cutaneous warts were collected for this study. There was no significant difference in the prevalence of warts between the genders (162 males and 167 females, respectively). The average age of the patients was 29.5 years and the age ranged from 5 to 67 years. Of the 329 patients, 166 patients (50.5%) were diagnosed common warts, 142 (43.2%) plantar warts, 5 (1.5%) flat warts and 15 (4.6%) were diagnosed of both common and plantar warts, one patient (0.3%) was diagnosed of both common and flat warts (Table 1).
The size of the generated HPV40 amplicon and HPV58 amplicon was 700 bp and 260 bp, respectively, and other 63 subtypes produced a band of 480 bp (Figure 1). Positive HPV DNA swabs were found in 315 of 345 warts. The rest of negative samples were then tested by β-globin primers to verify the validity of DNA samples. A total of 7 out of 30 negative HPV DNA samples showed negative β-globin amplification, indicating failure of clinical sampling and thus were excluded from the analysis. The final case series included 157 males and 165 females. The percentage of viral DNA detected by FAP primers was 93.2% (Table 1), with 95.1% in common wart, 89.2% in plantar wart and 100% in flat wart, and the combination of different kinds of cutaneous warts.
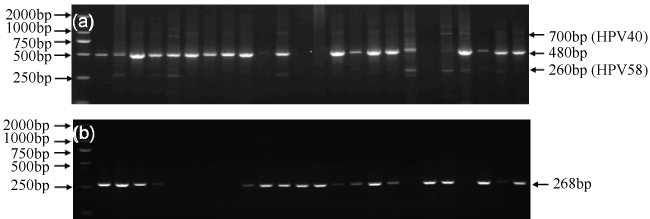 Figure 1: Polymerase chain reaction (PCR) results of representative cases. A) HPV-positive bands at 480 bp; 700 bp and 260 bp; B) β-globin at 268 bp.
View Figure 1
Figure 1: Polymerase chain reaction (PCR) results of representative cases. A) HPV-positive bands at 480 bp; 700 bp and 260 bp; B) β-globin at 268 bp.
View Figure 1
Table 1: Clinical information of the samples and the results of HPV detection in cutaneous warts. View Table 1
Cutaneous warts, caused by HPV, can affect a large broad range of the population worldwide, although the exact incidence remains unclear. HPV infection causes the proliferation of epidermal cells which is essential in the formation of skin lesions in any site of the body that severely influence patients' physical and psychological health. Cutaneous warts in general are diagnosed depending on their clinical features, but in some cases that are absent of typical manifestation, over- or under-diagnosis is commonly seen. Therefore, it is necessary to investigate an additional test to confirm the routine diagnosis. Culturing and propagation of virus in vitro are impractical and serological methods are lack of sensitivity [8]. However, simple and rapid HPV DNA test is favored in any clinical setting as a DNA sequencing of the PCR product can provide more accurate proof of the existence of HPV DNA. FAP primer pair-based method is a suitable option, because this method allows detection of 65 HPV subtypes with high sensitivity in a clinical setting, even the quantity of collected DNA is low. This method was originally introduced by Forslund in 1999 and has been successfully applied in many HPV typing studies [7], among a number of other primer sets used in clinical and epidemiology studies to detect HPV prevalence in verruca and healthy skin samples [9-15]. To our knowledge, this is the first study to evaluate HPV infection encompassing so many HPV types on a large cohort with cutaneous warts.
HPV DNA was detected in 93.2% of cutaneous warts, a rate which is higher than those reported in previous studies. This is probably because FAP primer set is capable of detecting multiple HPV types in each sample [16,17]. HPV DNA was undetectable in about 7% of the samples, but a negative result cannot completely rule out the presence of HPV infection. A possible explanation for the negative results could be attributed to the possibility that other related dermatosis was misdiagnosed as cutaneous warts, the virus load of HPV DNA was lower than the cutoff threshold point, or the primer set amplified PCR products have the limitation of covering all the HPV genotypes. However, there are limitations of this detection system that must be considered. First, swabbing the surface of a lesion may not accurately represent those present within biopsy tissue, due to ubiquitous presence of HPV, and a pre-taping may help to remove the contaminating virus. The presence of multiple HPV types on the surface of the warts are absent in the lower layers of tissue [18]. Second, although the use of the FAP primer set can improve the accuracy of HPV detection, there is still possibility of misdiagnosis because HPV may also be present in normal skin. Thirdly, our method was unable to detect most of the specific types of HPV as did by method based on the Luminex technology, etc.
In summary, we concluded that FAP primer set is a sensitive and effective method in a large number of swab samples clinically diagnosed with cutaneous warts, and this test method shows the benefits of convenience, low cost, more flexibility and high sensitivity and effectiveness for HPV detection. Moreover, this assay may be effective to distinguish between HPV-induced cutaneous wart and wart-like lesion that are not related to HPV infection. The technique can be an auxiliary examination of cutaneous warts. This method may also have the potential to determine HPV prevalence in larger series of skin carcinomas, benign lesions and normal skin of post-transplant population.
Such studies may improve our understanding of the relationship between HPV and the pathogenesis of these diseases.
We are grateful to all the participating patients from the First Hospital of China Medical University. Also, we are thankful for the doctors, Lanting Hu, Yali Gao, Yi Ren, Hanghang Jia, Zhengxiu Li, Juan Tang, and Tianci Shu for their contribution in the swabs collection. This work was partly supported by Public Welfare Project, Ministry of Health, China (201202013).