Journal of Infectious Diseases and Epidemiology
Development of Teicoplanin Tolerance by Staphylococcus epidermidis and Increased Susceptibility to Bacteriophage Type 92 by Methicillin-Resistant Staphylococcus aureus in Polymicrobial Biofilms
Jon M Riggs1, Robert JC McLean2*, Rodney E Rohde3 and Gary M Aron2
1Department of Microbiology and Molecular Biology, Oklahoma State University, USA
2Department of Biology, Texas State University, USA
3Clinical Laboratory Science Program, Texas State University, USA
*Corresponding author:
Robert JC McLean, Department of Biology, Texas State University, 601 University Drive, San Marcos, TX 78666, USA, Tel: 1-512-245-3365, Fax: 1-512-245-8713, E-mail: McLean@txstate.edu
J Infect Dis Epidemiol, JIDE-2-009, (Volume 2, Issue 1), Research Article; ISSN: 2474-3658
Received: February 01, 2016 | Accepted: February 26, 2016 | Published: February 29, 2016
Citation: Riggs JM, McLean RJC, Rohde RE, Aron GM (2016) Development of Teicoplanin Tolerance by Staphylococcus epidermidis and Increased Susceptibility to Bacteriophage Type 92 by Methicillin-Resistant Staphylococcus aureus in Polymicrobial Biofilms. J Infect Dis Epidemiol 2:009. 10.23937/2474-3658/1510009
Copyright: © 2016 Riggs JM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Staphylococcus epidermidis is a commensal commonly found in polymicrobial biofilms with Staphylococcus aureus. Given the increasing drug resistance in Staphylococci, we explored combination therapy with teicoplanin and bacteriophage type 92 (Siphoviridae) on the survival and antibiotic tolerance of methicillin-resistant S. aureus (MRSA) and S. epidermidis to teicoplanin and bacteriophage in 48-h pure and mixed culture biofilms. The combination of teicoplanin and bacteriophage was more effective against MRSA, but not S. epidermidis monocultures, than the use of either teicoplanin or phage alone. In polymicrobial biofilms, however, MRSA acquired increased susceptibility to phage infection and S. epidermidis acquired increased tolerance to teicoplanin, as well as increased fitness. The results demonstrate bacteriophage alone was more effective against MRSA and S. epidermidis in polymicrobial biofilms compared to the combination of bacteriophage and teicoplanin.
Keywords
Biofilms, Bacteriophage, Antibiotic, Tolerance, MRSA, Mixed culture
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) has emerged as one of the most dangerous and widely publicized pathogens in the United States since its initial identification [1,2]. Originally limited to nosocomial, or healthcare-associated infections (HA-MRSA), MRSA has since been identified as a serious public health threat to persons with no affiliation with hospital settings [3]. In the 2000s, MRSA emerged in community settings [4] including USA college campuses associated with clinical programs resulting in the distinction between HA-MRSA and community-acquired MRSA (CA-MRSA) [5]. The gene responsible for methicillin resistance in S. aureus, mecA, is not found on methicillin-susceptible strains of S. aureus [4]. A number of infections are associated with MRSA including those associated with medical devices, soft tissue abscess infections and occasionally more invasive conditions. MRSA infections are often polymicrobial and frequently include Staphylococcus epidermidis [6]. In medical device infections, MRSA often grows as surface-adherent biofilm communities. Biofilm growth results in increased antimicrobial resistance, particularly in a polymicrobial community [7].
Beta-lactam resistant staphylococcal infections are often treated with glycopeptide antibiotics, such as vancomycin and teicoplanin [8]. While similar to vancomycin in bactericidal spectrum, teicoplanin is not currently FDA-approved for use in the United States [8]. Outside the US, several studies have investigated the efficacy and potential side effects of teicoplanin in comparison to other glycopeptide antibiotics including vancomycin [9]. Since staphylococcal species easily attain antibiotic resistance, and some bacteriophages are known to degrade biofilm matrices, attention has refocused on the therapeutic use of bacteriophages [10].
Bacteriophages have been used since the earliest 20th century to treat a number of infections [11,12]. In Europe and North America phage therapy has been largely supplanted by antibiotic therapy [11] although it has still been pursued in the former Soviet Union and other eastern countries [13]. With the increasing resistance of bacterial infections to antibiotics, phage therapy is being reexamined. In this context, examples of experimental strategies include the use of phage cocktails targeting multiple bacterial strains [14] and combination phage-antibiotic therapy [15]. A number of phage depolymerase enzymes have been described, which enable phage to penetrate biofilm matrix polymers [10,16]. While these work well in monoculture biofilms, polymicrobial biofilms have variable matrix chemistries and our previous study [17] has shown matrix polymers from a non-target host organism to be capable of interfering with some phage. Here, we investigate the effectiveness of teicoplanin and bacteriophage combination therapy in monoculture and polymicrobial biofilms of methicillin-resistant S. aureus and methicillin-susceptible S. epidermidis. These two organisms are a frequent cause and commonly co-exist in transcutaneous medical device infections [6,18].
Materials and Methods
Bacterial strains, bacteriophage, media, and culture conditions
Methicillin-resistant Staphylococcus aureus (ATCC 37741), Staphylococcus epidermidis (ATCC 12228), and Bacteriophage type 92 (ATCC 33741-B) [19] were obtained from the American Type Culture Collection (Manassas, VA). S. aureus and S. epidermidis were grown in Tryptic Soy broth (TSB) (Accumedia Inc., Lansing, Michigan) at 37°C in an orbital rotating shaker water bath. Bacteriophage type 92 was used to infect S. aureus and S. epidermidis. Bacteriophage 92 stocks were prepared by infecting early log phase S. aureus using the agar overlay technique as previously described [20]. Briefly, 350 μL of early log phase S. aureus was added to 3.5 mL of 0.38% (w/v) agar in TSB. 100 μL of 106 (plaque-forming units (PFU)/ml) phage was added to yield confluent lysis. Following 24 h at 37°C, phage was eluted from the agar overlay in 5 mL TSB at 4°C. The phage-containing liquid was centrifuged at 4°C for 20 min at 4000g (Eppendorf Centrifuge model 5810R, Hamburg, Germany). Supernatant was filtered (0.45 μm) and phage titers (PFU/ml) were determined by a soft-agar overlay assay [20]. Teicoplanin (Sigma-Aldrich Co. St Louis, MO) stocks were prepared in deionized water followed by filter sterilization. Teicoplanin at a concentration of 10 μg/mL was used for all experiments. During dilution plating of mixed cultures, S. aureus and S. epidermidis could be easily distinguished by growth on mannitol salt agar. A minimum of three biological replicates was performed for each study.
Biofilm formation
Overnight cultures (18 h) of S. aureus or S. epidermidis were diluted 1:500 and 200 μL was dispensed into clear, sterile, non-tissue treated 96-well plates (Fisher Scientific Co., Horizon Ridge, CT). For polymicrobial biofilms, 50 μL of each overnight culture were diluted in 50 mL TSB and 200 μL of the diluted culture was dispensed into plates. Plates were incubated for 48 h at 37°C with rocking at 10°C, at 15 rpm (VWR Signature™ Incubating Rocking Platform Shaker, VWR, Houston, TX).
Biofilm bacteriophage and antibiotic susceptibility assays
Supernatant was removed from 48 h S. aureus biofilms and biofilms washed with 200 μL PBS. Biofilms were treated with either phage (multiplicity of infection (MOI) 10), teicoplanin (10 μg/mL), or a combination of both contained in 200 μL TSB at 37°C. After 12h treatment, biofilms were sonicated for 1 min at 40 kHz, and colony forming units (CFU) determined by dilution plating on TSA plates.
Determination of increase in tolerance to teicoplanin by S. epidermidis
Susceptibility of S. epidermidis to teicoplanin was determined using the Kirby-Bauer disk diffusion technique, as described by the Clinical and Laboratory Standards Institute [21] with TSA substituted for Mueller-Hinton agar. S. epidermidis susceptibility to teicoplanin was tested before and after exposure to S. aureus in polymicrobial biofilms. S. epidermidis recovered from polymicrobial biofilms was sub-cultured nine successive times and each sub-culture was tested for susceptibility to teicoplanin.
Effect of S. epidermidis supernatant on S. aureus phage sensitivity
S. aureus sensitivity to phage was determined by growing S. aureus biofilms with 100 μL of S. epidermidis supernatant for 48 h at 37°C. Culture supernatant was obtained after 18 h planktonic culture from either S. epidermidis monoculture, or a mixed S. epidermidis –MRSA culture. Supernatants were prepared by centrifugation (3000 g for 20 min) followed by filtration through a 0.45 μm pore-size filter. S. aureus biofilms were washed with 200 μL PBS and infected with phage at MOI 10. Biofilms were enumerated by sonication and dilution plating as described above.
Transmission electron microscopy (TEM)
For TEM examination, 5 μL of 1010 PFU/mL phage were placed on 200 mesh count, Formvar-coated copper grids (Electron Microscopy Sciences, Hatfield, PA) overnight at room temperature. Samples were stained with 2% uranyl acetate for one minute and viewed using a JEM-1200 EXII at an accelerating voltage of 60 KV.
Results
Phage type 92 characterization
Phage type 92 was originally isolated in 1979 from MRSA isolates [19]. TEM examination of phage 92 (Figure 1), revealed icosahedral capsids approximately 62 nm in diameter and long, flexible tails approximately 175 nm in length. On the basis of viral morphology and established taxonomy guidelines [22], phage 92 belongs to the family Siphoviridae.
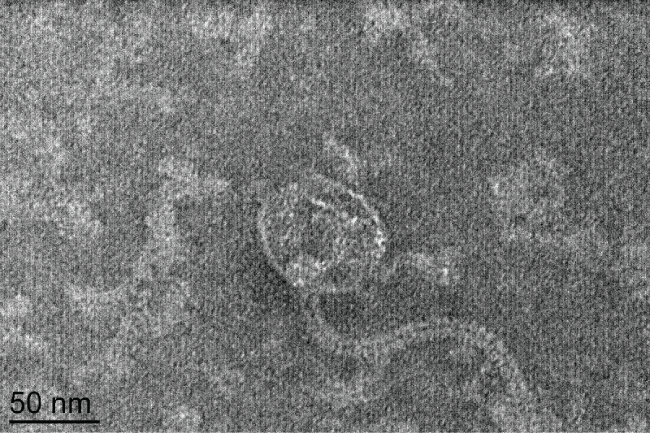
.
Figure 1: TEM micrograph of Phage 92. Based on the flexible tail and icosahedral head [22], this phage should be classified in the family Siphoviridae.
View Figure 1
Effect of bacteriophage and teicoplanin, on monoculture and mixed culture biofilms
Growth of S. aureus and S. epidermidis following treatment with phage 92, teicoplanin and combination treatment is shown in Figure 2. We also calculated changes in fitness (expressed as a ratio of (log CFU treated culture/log CFU monoculture control)) which is shown in Figure 3. When compared to untreated controls, the most effective treatment of monoculture MRSA biofilm cultures was combination phage and teicoplanin. Since phage require growing host cells in order to replicate, the enhanced success of the combination treatment suggests that teicoplanin did not completely block S. aureus growth when added in combination with phage 92. Different results were seen during treatment of mixed culture S. aureus. Here, the most effective treatment was phage alone. One likely explanation for this phenomenon is that phage attack on the host S. aureus facilitated competition from S. epidermidis, which was minimally affected by the phage during monoculture growth. Of interest, growth in mixed culture resulted in less impact of teicoplanin against MRSA alone or in combination with phage 92.
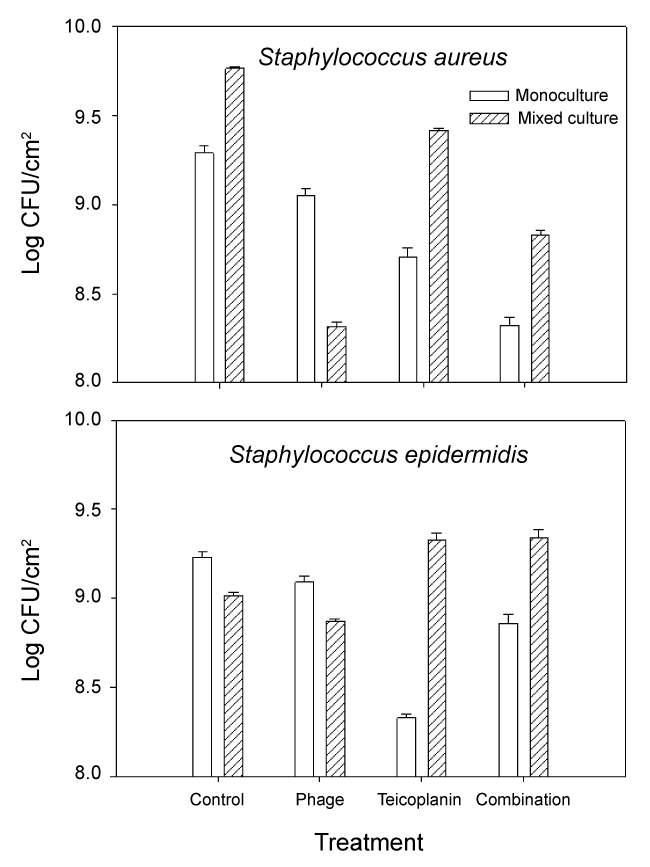
.
Figure 2: Growth of MRSA (S. aureus) and S. epidermidis biofilms following exposure to phage 92, teicoplanin, or combination phage 92 and teicoplanin. Details are in the text.
View Figure 2
In monoculture S. epidermidis cultures, the most effective treatment was teicoplanin. This is not surprising as teicoplanin has been reported to be effective against this organism [9]. In contrast, mixed culture results were quite different. In untreated mixed-culture biofilms, MRSA outcompeted S. epidermidis which is reflected both in culture numbers (Figure 2 control) and fitness calculations (Figure 3 control). Although MRSA is the host organism for phage 92, there was a slight decrease in S. epidermidis numbers and fitness (P < 0.001) relative to the mixed culture control, suggesting that some phage influence or competition from MRSA was occurring in the presence of phage. In contrast to the reduced fitness seen in S. epidermidis monocultures with teicoplanin alone or in combination with phage 92; the fitness of S. epidermidis was significantly increased in mixed culture (P < 0.001 in comparison to monoculture results) when grown in mixed culture. We also observed that repeated subculture of S. epidermidis with MRSA decreased teicoplanin susceptibility and the onset of resistance (Figure 4).
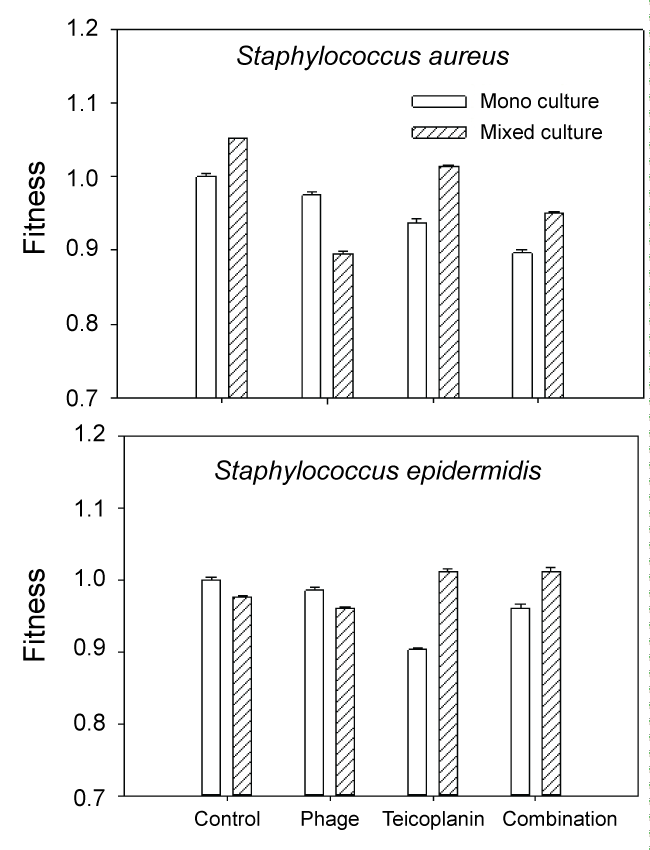
.
Figure 3: Fitness of S. aureus and S. epidermidis expressed as the ratio of log CFU treated culture/log CFU untreated (control) monoculture.
View Figure 3
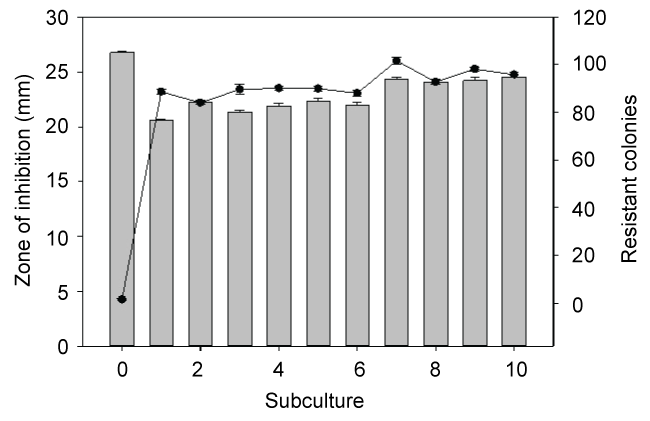
.
Figure 4: Subculture of S. epidermidis with MRSA resulted in a slight increase of tolerance as reflected in a decrease of the zone of inhibition (bar graph). As well the number of resistant colonies (line graph) increased. The notable changes occurred after the organisms were initially grown together (subculture 1), with minimal changes seen during subsequent subcultures.
View Figure 4
Discussion
The decreased antibiotic susceptibility of monoculture biofilms has been known for over 30 years [23] and a number of mechanisms for biofilm-mediated resistance have been described including altered growth rates within biofilms, biofilm-specific resistance genes and the presence of dormant persister subpopulations (reviewed in [24,25]). Biofilms including MRSA wound infections [6] typically contain polymicrobial communities and there is increasing interest in studying antimicrobial susceptibility in that context. Two studies showed that increased bacterial community diversity resulted in higher biofilm cell density [26,27]. In one of these studies, increased biofilm resistance to disinfection was due to general interactions among all community members [26]. In the second study, biofilm resistance was entirely due to the presence of a single resistant community member [27]. Combination phage-antibiotic treatments have shown promise in the treatment of monoculture biofilms e.g. [28,29], however their effectiveness against mixed cultures is less clear. With planktonic cultures, Harcombe and Bull [30] showed that phage could enhance microbial competition in a two-species system by targeting one component organism. However, biofilm growth does enable many bacteria to persist in spite of competition [31].
Our data would suggest that microbial competition does play a role in the efficacy of some antimicrobial treatments against some biofilms, most notably the phage 92 treatment of MRSA in mixed cultures (Figure 3). Competition was not evident during treatment of mixed culture biofilms with teicoplanin alone or in combination with phage 92. The increased fitness of both MRSA and S. epidermidis during teicoplanin treatment in comparison to the monoculture results (P < 0.001) suggests that the two normally competitive organisms, exhibit synergy (facilitation) in the presence of teicoplanin. In previous work with environmental isolates, we found that the presence of one disinfectant (betadine) resistant organism could confer resistance to an entire mixed biofilm population [27] likely through facilitation, so this finding is not unprecedented.
In planktonic monoculture or mixed Pseudomonas aeruginosa and Escherichia coli cultures, we observed that the administration of sub-lethal concentrations of antibiotics or phage is followed rapidly by the onset of resistance through the planktonic populations likely through mutation and natural selection. In contrast, while biofilm populations of these organisms were protected by this mode of growth, a significant proportion of organisms within biofilms retained susceptibility to phage or antibiotics when released from biofilms by sonication, diluted and cultured on antibiotic- or phage-containing agar [15,17]. We tested the influence of previous co-culture growth on the teicoplanin susceptibility of S. epidermidis using a disk diffusion assay. We also measured the number of spontaneous teicoplanin-resistant colonies which grew within the zone of inhibition. During these studies, S. epidermidis was sub-cultured overnight up to ten times with MRSA and then purified and tested. As seen in Figure 4, there was a slight but significant (P < 0.01) decrease of susceptibility following the first sub-culture. In subsequent subcultures, while there was some variability in the zone of inhibition, it was still significantly less than the original culture (designated as subculture 0) before mixed culture growth. Similarly, there was an increase in the number of resistant colonies seen after the first sub-culture with the numbers increasing from 4 to > 90 in most cases. Although S. aureus (ATCC 37741) is resistant to methicillin, it is sensitive to teicoplanin, which would rule out horizontal gene transfer of a pre-existing resistance gene to S. epidermidis. Although we do not have a direct explanation for this phenomenon, it is possible that mixed culture growth of S. epidermidis with S. aureus activated mechanisms that enabled S. epidermidis to better tolerate exposure to teicoplanin. Further work will be needed to elucidate the mechanism(s) behind this phenomenon.
Conclusions
Many S. aureus infections involve polymicrobial interactions with S. epidermidis and biofilm growth [6]. In the current study, interactions between S. aureus and S. epidermidis strains used affected the efficacy of teicoplanin antibiotic and phage treatment. In some cases, when one organism was targeted (MRSA specific phage 92 in mixed culture), mixed culture interactions enhanced the treatment due to microbial competition, whereas in other cases in which both organisms were targeted by teicoplanin, the efficacy was reduced possibly due to facilitation interactions. Overall these data emphasize the importance of studying mechanisms of antimicrobial treatments in a polymicrobial biofilm growth system.
Acknowledgments
This work was funded by the Homer E. Prince Microbiology Endowment to GM Aron. We thank Alissa Savage for help with TEM.
Conflict of Interest
The authors declare no conflict of interest.
References
-
Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, et al. (2007) Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298: 1763-1771.
-
Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, et al. (2001) Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357: 1225-1240.
-
Ubukata K, Nonoguchi R, Matsuhashi M, Konno M (1989) Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J Bacteriol 171: 2882-2885.
-
Ma XX, Ito T, Tiensasitorn C, Jamklang M, Chongtrakool P, et al. (2002) Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother 46: 1147-1152.
-
Rohde RE, Denham R, Brannon A (2009) Methicillin resistant Staphylococcus aureus: carriage rates and characterization of students in a Texas university. Clinical Laboratory Science 22: 176-184.
-
Peel TN, Cheng AC, Buising KL, Choong PF (2012) Microbiological aetiology, epidemiology, and clinical profile of prosthetic joint infections: are current antibiotic prophylaxis guidelines effective? Antimicrob Agents Chemother 56: 2386-2391.
-
Pozzi C, Waters EM, Rudkin JK, Schaeffer CR, Lohan AJ, et al. (2012) Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog 8: e1002626.
-
Svetitsky S, Leibovici L, Paul M (2009) Comparative efficacy and safety of vancomycin versus teicoplanin: systematic review and meta-analysis. Antimicrob Agents Chemother 53: 4069-4079.
-
Gallon O, Guillet-Caruba C, Lamy B, Laurent F, Doucet-Populaire F, et al. (2009) In vitro activity of daptomycin against Staphylococci isolated from bacteremia and community-onset skin and soft tissue infections in France: data from two nationwide studies. Eur J Clin Microbiol Infect Dis 28: 1209-1215.
-
Parasion S, Kwiatek M, Gryko R, Mizak L, Malm A (2014) Bacteriophages as an alternative strategy for fighting biofilm development. Pol J Microbiol 63: 137-145.
-
Hanlon GW (2007) Bacteriophages: an appraisal of their role in the treatment of bacterial infections. Int J Antimicrob Agents 30: 118-128.
-
d'Herelle F (1931) Bacteriophage as a Treatment in Acute Medical and Surgical Infections. Bull N Y Acad Med 7: 329-348.
-
Marcuk LM, Nikiforov VN, Scerbak JF, Levitov TA, Kotljarova RI, et al. (1971) Clinical studies of the use of bacteriophage in the treatment of cholera. Bull World Health Organ 45: 77-83.
-
Krylov VN (2014) Bacteriophages of Pseudomonas aeruginosa: long-term prospects for use in phage therapy. Adv Virus Res 88: 227-278.
-
Coulter LB, McLean RJ, Rohde RE, Aron GM (2014) Effect of bacteriophage infection in combination with tobramycin on the emergence of resistance in Escherichia coli and Pseudomonas aeruginosa biofilms. Viruses 6: 3778-3786.
-
Sutherland IW, Hughes KA, Skillman LC, Tait K (2004) The interaction of phage and biofilms. FEMS Microbiol Lett 232: 1-6.
-
Kay MK, Erwin TC, McLean RJ, Aron GM (2011) Bacteriophage ecology in Escherichia coli and Pseudomonas aeruginosa mixed-biofilm communities. Appl Environ Microbiol 77: 821-829.
-
Krimmer V, Merkert H, von Eiff C, Frosch M, Eulert J, et al. (1999) Detection of Staphylococcus aureus and Staphylococcus epidermidis in clinical samples by 16S rRNA-directed in situ hybridization. J Clin Microbiol 37: 2667-2673.
-
Schaefler S, Perry W, Jones D (1979) Methicillin-resistant strains of Staphylococcus aureus phage type 92. Antimicrob Agents Chemother 15: 74-80.
-
Adams MH (1959) Methods of study of bacterial viruses, Bacteriophages, New York: Interscience Publications.
-
Li G, Ma X, Deng L, Zhao X, Wei Y, et al. (2015) Fresh Garlic Extract Enhances the Antimicrobial Activities of Antibiotics on Resistant Strains in Vitro. Jundishapur J Microbiol 8: e14814.
-
Ackermann HW (2009) Phage classification and characterization. Methods Mol Biol 501: 127-140.
-
Nickel JC, Ruseska I, Wright JB, Costerton JW (1985) Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob Agents Chemother 27: 619-624.
-
Lewis K (2007) Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5: 48-56.
-
Zhang L, Fritsch M, Hammond L, Landreville R, Slatculescu C, et al. (2013) Identification of genes involved in Pseudomonas aeruginosa biofilm-specific resistance to antibiotics. PLoS One 8: e61625.
-
Burmølle M, Webb JS, Rao D, Hansen LH, Sørensen SJ, et al. (2006) Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol 72: 3916-3923.
-
Whiteley M, Ott JR, Weaver EA, McLean RJC (2001) Effects of community composition and growth rate on aquifer biofilm bacteria and their susceptibility to betadine disinfection. Environ Microbiol 3: 43-52.
-
Bedi MS, Verma V, Chhibber S (2009) Amoxicillin and specific bacteriophage can be used together for eradication of biofilm of Klebsiella pneumoniae B5055. World J Microbiol Biotechnol 25: 1145-1151.
-
Fu W, Forster T, Mayer O, Curtin JJ, Lehman SM, et al. (2010) Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob Agents Chemother 54: 397-404.
-
Harcombe WR, Bull JJ (2005) Impact of phages on two-species bacterial communities. Appl Environ Microbiol 71: 5254-5259.
-
Elias S, Banin E (2012) Multi-species biofilms: living with friendly neighbors. FEMS Microbiol Rev 36: 990-1004.





