A supramolecular indicator system (cellulose-nanodiamonds-urease) for reusable biochemical detection of urea has been fabricated using sequential immobilization of the components. Modified nanodiamonds (MNDs) were covalently immobilized onto DEAE cellulose granules via the nucleophilic addition reaction. At DEAE cellulose: MND ratio of 4:1 (w/w), up to 110 µg of nanoparticles bound onto 1 mg of the polymer during the addition reaction. Urease was immobilized by covalent conjugation onto the polymer-MND composite with the benzoquinone-activated surface. In comparative experiments, the enzyme was immobilized onto initial polymer granules via nonspecific adsorption and covalent conjugation. However, when these indicator systems were repeatedly used to detect the analyte, the enzyme was considerably inactivated, and that was evidenced by a decrease in the colored product yield. At the same time, the enzyme covalently bound onto the DEAE cellulose-MND composite showed higher functional efficacy and enabled more stable yields of the colored product in repeated urea assays. Comparative experiments with the indicator systems repeatedly used to detect urea at 37 ºC demonstrated that the enzyme covalently conjugated onto the DEAE cellulose-MND composite showed greater thermo stability, and its activity was reduced at a much slower rate than the activity of the enzyme covalently bound to the polymer. The data obtained in the present study offer the prospect of designing a new type of reusable indicator assay systems (polymer carrier-nanodiamond-biomarker supramolecular systems) for biomedical analytical applications.
Nanodiamond, Cellulose, Composite material, Urease, Indicator system, Urea detection
Nowadays researchers around the world are intensively developing a great number of various types of analytical tools and biosensors such as enzyme-based, tissue-based, DNA biosensors, immunosensors, thermal and piezoelectric biosensors. Biosensors have been applied in many fields such as food industry, fermentation industry, medical field, plant biology sector, marine sector etc. [1-5], because they provide better stability and sensitivity as compared with the traditional methods.
In particular, the efficiency and selectivity of enzymatic catalysis is of great importance in biomedical applications and clinical diagnostics. However, free enzymes have some limits including the loss of catalytic activity after one cycle, low stability, and low activity in the organic phase. They also cannot be separated from the substrate or product [6,7]. The immobilization of enzymes on the carrier can overcome these disadvantages efficiently. Mainly, there are three kinds of immobilization: binding with a support (adsorption, ionic binding, and covalent binding), cross-linking, and entrapment [6,8]. The covalent binding is a practical method for enzyme immobilization: the protection effect of the carrier can enhance the stability of the enzyme, and the leakage of the enzyme can be reduced by increasing the strength of the bonds [9].
Applications of nanomaterials have extended into the field of biomedical sensing expanding the arsenal of currently employed techniques. In particular, nanoparticles of various physical and chemical origins can be an attractive material for developing indicator and diagnostic assay systems as a carrier for immobilizing peptide biomarkers (including enzymes) [10-15]. Along with the fact that a high surface-to-volume ratio offered by nanoparticles provides a significantly higher concentration of the immobilized enzymes than that afforded by immobilization on planar 2D surfaces, the integration of nanoparticles into enzyme carrier structures maintain or even enhance immobilized enzyme performance [16-18].
In this regard, detonation nanodiamond [19,20] are well explored for biomedicine related issues due to their chemical stability, large specific area, facile surface tailoring for specific applications, high adsorption capacity and non-toxicity [21-24]. Along with a high colloidal stability in dispersion media, these properties of modified nanodiamonds (MND) provide for their potential use as a basis for a new class of bioanalytical devices for targeting and quantifying analytes of interest [25-27]. The technology of MND obtaining includes chemical modification of the surface of initial commercial nanodiamonds by NaCl treatment, that reduces the number of inorganic admixtures, e.g. metal ions [24,28]. MND are characterized by high sedimentation stability in aqueous suspensions (hydrosols) and can be separated by differential centrifuging into narrow fractions with various clusters size (nanoparticle aggregates).
In our previous studies, we showed that MNDs can be used to construct systems for biochemical diagnostics and targeted delivery of drugs and new antibacterial preparations [29-31]. The MND-enzyme systems were used in biochemical assays of physiologically significant analytes [29]. The immobilization of the sensing element (nanoparticle-biomarker conjugate) on a solid support can be adapted and advanced the experimental indicator system for clinical analytical applications. The purpose of this study was to construct and test a biochemical assay system for detecting urea in order to provide an experimental basis for designing a reusable indicator systems based on the cellulose-nanodiamond-biomarker supramolecular complexes.
The objective of this work was to construct and test a model biochemical assay system for detecting urea in order to provide an experimental basis for designing a reusable indicator systems based on the cellulose-nanodiamond-biomarker supramolecular complexes.
The polymeric carrier (support) for the indicator system was prepared from diethylaminoethyl (DEAE) cellulose granules (Serva, Germany). A sample of DEAE cellulose granules was mixed with deionized (DI) water, prepared using a Milli-Q system (Millipore, U.S.). The suspension was incubated for 12 h at a temperature of 20-22 ºC to allow the granules to swell. Then, DI water was decanted. The polymeric granules were washed with DI water thrice to remove residual preservatives contained in the commercial DEAE cellulose.
The experiments were conducted using MND with high colloid resistance in hydrosols and average cluster size (nanoparticle aggregates) d50 = 50 nm. Size distribution of MND clusters in hydrosols was measured with a Zetasizer Nano ZS (Malvern Instruments Ltd., U.K.). MND were obtained from commercial nanodiamonds (LTD "Real-Dzerzhinsk", Russia) by chemical modification using original technique described previously [28]. The technique consisted of several stages. First, nanoparticle hydrosol was obtained from nanodiamond powder and deionized water. A modifying agent (NaCl) was added to the obtained suspension; the suspension was mixed and allowed to stay at room temperature for 1 h until precipitation of all particles. The supernatant was removed by decantation; the sediment of nanoparticles was washed with deionized water to remove the agent. After washing hydrosol with MND was differentially centrifuged at various forces (centrifuge Avanti J-E, Beckman Coulter) to isolate MND fractions with different cluster sizes (nanoparticle aggregates). The obtained fraction of MND was exsiccated to obtain dry substance of nanoparticles. We used the hydrosol of MND (1% wt), which was prepared by adding deionized water to the sample of nanoparticles powder.
Urease (EC 3.5.1.5 from jack beans) was purchased from Vektor-Best, Russia. The reagents used in this study to determine urea in blood serum and urine were from the Novokarb kit (Vektor-Best, Russia). The kit contained a urea calibrator (analyte concentration of 5 mmol/L), and solutions of chemical reagents (sodium salicylate, nitroprusside, hypochlorite, and hydroxide). Immobilization was performed using an aqueous solution of urease, which had been prepared by dialyzing the initial enzyme solution against DI water, by ultrafiltration through a 30 kDa cutoff membrane (EMD Millipore Amicon, Germany).
The polymeric carrier-nanodiamond-biomarker indicator system was fabricated by sequential immobilization of the components (Figure 1a). The DEAE cellulose-MND composite was prepared using the nucleophilic addition reaction [32]. The aqueous suspension of cellulose granules was mixed with MND hydrogel at a ratio of 4:1 (polymer: nanoparticles, w/w). Hydrochloric acid (Reakhim, Russia) was added to the mixture to reach the final concentration of 5 mM, and the mixture was incubated at a temperature of 20-22 ºC for 1 h under continuous agitation at 180 rpm (Orbital Shaker OS-10, BIOSAN, Latvia). Then, the polymer-nanoparticle composite was collected by centrifugation (Centrifuge 5415R, Eppendorf, Germany) at 2300 g for 5 min at 20 ºC. The settled polymer was washed repeatedly (3 times with DI water, 1 time with a 1 M NaCl solution, and 3 times with DI water) to remove MND particles that remained unbound to the DEAE cellulose granules. Each time, the granules were resuspended in the fresh washing solution and collected by centrifugation as described above. In every phase (after the nucleophilic addition reaction and washing), the supernatants were collected for spectral analysis (an UV-1800 spectrophotometer, Shimadzu, Japan) for the presence of MNDs. The amount of the MNDs unbound to the polymer was determined from the optical density of the initial MND hydrosol and supernatant at a wavelength of 400 nm.
 Figure 1: A schematic representation of a) Constructing supramolecular indicator system (DEAE cellulose-MND- urease); b) Determining the activity of the enzyme in the experimental supramolecular system. View Figure 1
Figure 1: A schematic representation of a) Constructing supramolecular indicator system (DEAE cellulose-MND- urease); b) Determining the activity of the enzyme in the experimental supramolecular system. View Figure 1
Urease was immobilized on the polymeric granules through nonspecific adsorption and covalent bonding. Covalent bonding was used to immobilize the enzyme onto the DEAE cellulose-MND composite. For the enzyme adsorption, the urease solution was added to the polymeric granule suspension, and the mixture was incubated at a temperature of 20-22 ºC for 1 h under continuous agitation at 180 rpm (Orbital Shaker OS-10). Then, the polymeric granules with the enzyme adsorbed on them were collected by centrifugation (Centrifuge 5415R) under the conditions described in the previous section. The sediment was washed with DI water thrice to remove the residual enzyme, which remained unbound to the granules; each time, the sediment was resuspended in a fresh portion of water and collected by centrifugation. After washing, the granules with the adsorbed urease were resuspended in DI water and used in the investigation. Urease was covalently bonded onto the granules of the polymer and the composite, whose surfaces had been preliminarily activated by benzoquinone (Figure 1a) using the conventional procedure [33-35]. Due to this pretreatment, the subsequent bonding of the proteins (including enzymes) onto the activated support is conducted under relatively mild conditions, preventing protein denaturation [35]. The surface of the cellulose and DEAE cellulose-MND composite granules was activated with benzoquinone, and urease was bonded as described in detail in our previous studies, which were devoted to the construction of the systems for targeted delivery of bioactive substances and biochemical diagnostics based on MNDs and various proteins and enzymes [29,30]. After covalent immobilization of urease, the polymeric and composite granules were collected by centrifugation and washed repeatedly (3 times with a 0.25 M NaCl solution and 1 time with DI water) to remove the enzyme unbound to the support. After washing, the granules with the covalently bonded urease were resuspended in DI water and used in experiments.
The activity of the enzyme in the experimental supramolecular systems was determined using the urease salicylate-hypochlorite method (modified Berthelot reaction) [36], which yield a colored reaction product in the urea assay. The method consists of two stages (Figure 1b) the enzymatic and the non-enzymatic (chemical) stages. In stage 1, urea is hydrolyzed by urease to form ammonia, which, in stage 2, interacts under alkaline conditions with the chemical components of the reaction mixture to yield a colored product. In the urea assay using the experimental indicator systems, the enzymatic stage was separated from the non-enzymatic one in order to prevent inactivation of the immobilized enzyme by chemical reagents (chiefly sodium hypochlorite and hydroxide) and to prepare reusable indicator systems for determining the analyte. The preliminary experiments showed that even a single run of both reactions in the presence of the indicator systems completely inactivated the immobilized enzyme. In the assay of urease activity, the reaction mixture of a total volume of 250 µl contained 145 µl DI water, 100 µl aqueous suspension of the indicator system (amount of the immobilized enzyme was 30 µg in the reaction), and 5 µl urea calibrator (with analyte concentration in the sample 0.1 mmol/L). All components were mixed and incubated for 5 min at a temperature of 20-22 ºC; then, the systems were collected by centrifugation. The supernatants were collected and mixed with the solutions of the chemical reagents for the non-enzymatic stage, which had been prepared in situ in DI water following the instructions of Vektor-Best. The samples were incubated for 5 min at the temperature mentioned above. The amount of the colored product was determined spectrophotometrically (UV-1800) by measuring the optical density of the samples at a wavelength of 700 nm. The indicator systems intended for reuse were collected by centrifugation after the enzymatic reaction. The supernatants were collected for conducting the chemical reaction and for the spectral determination of the colored product. The sediment was washed twice (1 time with a 125 mM NaCl solution and 1 time with DI water) to remove the residual substrate (urea) and product of the enzymatic reaction (ammonia). Then, the systems were resuspended in DI water, and the analyte was added to start the enzymatic reaction. The thermo stability of the enzyme on the reused indicator systems was estimated by incubating the samples in a thermostat (Thermo bath TB-85, Shimadzu, Japan) at 37 ºC during the enzymatic reaction.
Each measuring experiment was performed in triplicate. Error bars were generated as a standard deviation of the mean from 3 replicates.
The microscopic examination of the experimental supramolecular system was conducted using an AxioImager M2 light microscope (Carl Zeiss, Germany). To visualize the immobilized enzyme, it was labeled with the FITC (fluoresce in isothiocyanate) fluorescent dye (Sigma, U.S.) prepared in dimethyl sulfoxide.
In the first phase of fabricating the supramolecular system for determining urea, we attempted to construct a composite based on cellulose granules and MNDs as a support for immobilization of the biomarker-urease. To obtain the DEAE cellulose-MND composite, we used the nucleophilic addition reaction (Figure 2).
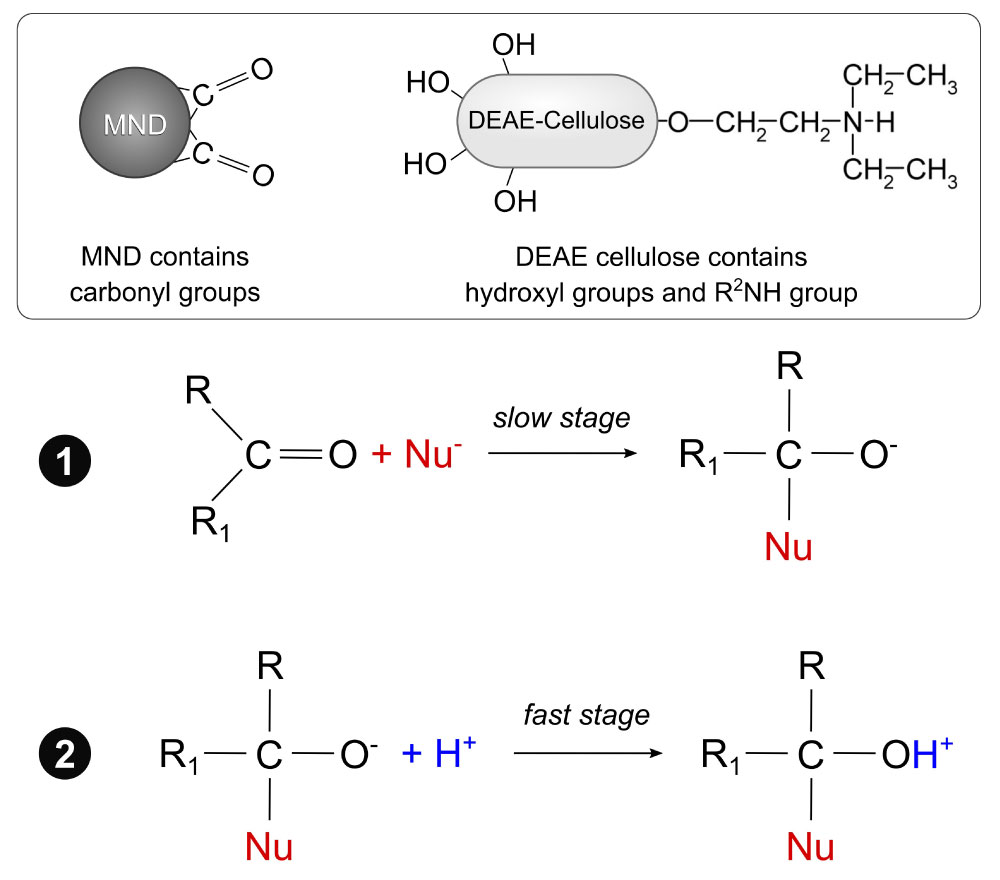 Figure 2: Schematic expression of the nucleophilic addition reaction between MND and DEAE cellulose via the most common two-step bimolecular mechanism AdN2: Slow attachment of the nucleophile by multiple bonds with the formation of a carbanion, which is then quickly attacked by an electrophilic particle. View Figure 2
Figure 2: Schematic expression of the nucleophilic addition reaction between MND and DEAE cellulose via the most common two-step bimolecular mechanism AdN2: Slow attachment of the nucleophile by multiple bonds with the formation of a carbanion, which is then quickly attacked by an electrophilic particle. View Figure 2
The choice of this reaction was based on the following reasons. The reactions of addition via the most common bimolecular mechanism AdN2 (forming a C=O bond) are a two-step process, in which nucleophiles may be represented by OH, R2O, NH2, R2NH, R2CH, etc. [32]. As reported in a previous study [27], there are a large number of carbonyl groups on the MND surface. Cellulose is a homopolymer made up of D-glucose residues, and it contains a large number of hydroxyl groups [37]. Moreover, DEAE cellulose contains R2NH. Since the addition reactions with oxygen nucleophiles are catalyzed by acids [32], the DEAE cellulose-MND composite was prepared by incubating the components in DI water containing 5 mM HCl.
After optimizing the reaction conditions (component ratio, suspension agitation speed, and time and temperature of incubation), we managed to produce a DEAE cellulose-MND composite using the nucleophilic addition method. That was evidenced by a change in the appearance of the polymeric granules after the reaction (Figure 3). The initially milky-white polymer granules turned brown because of the MNDs bound to them during the acidic reaction.
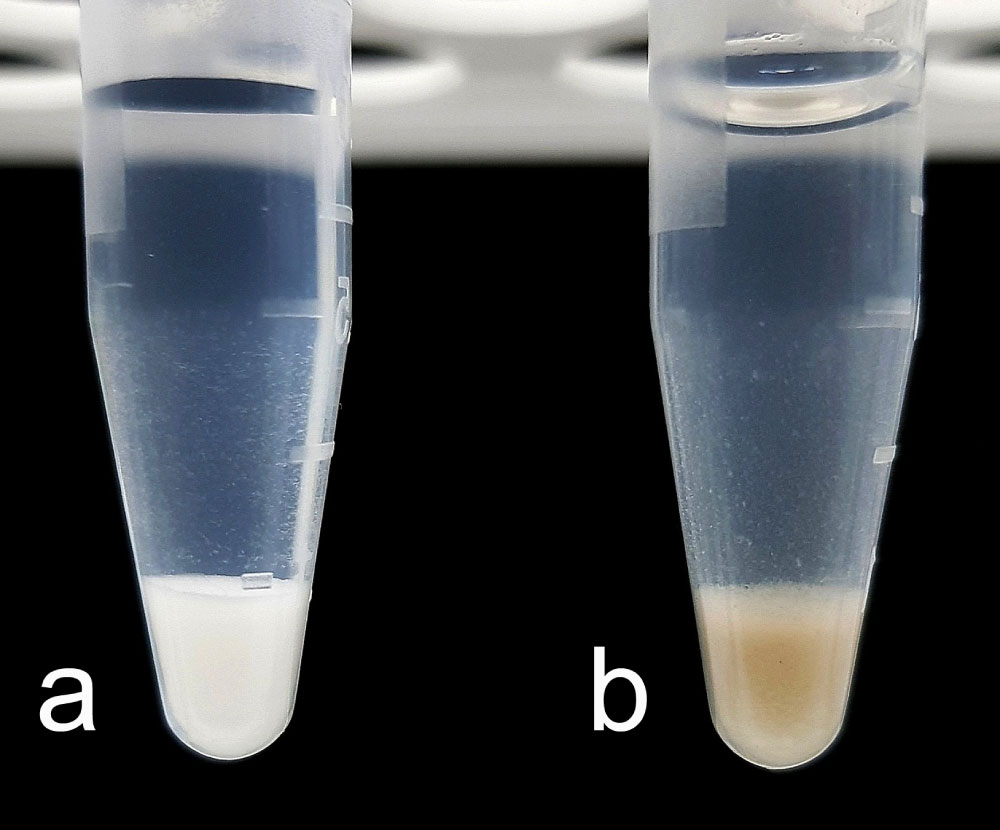 Figure 3: a) Initial DEAE cellulose granules b) And the DEAE cellulose-MND composite. View Figure 3
Figure 3: a) Initial DEAE cellulose granules b) And the DEAE cellulose-MND composite. View Figure 3
The amount of the MNDs immobilized on the polymer was calculated using the data of the spectral analysis of the initial MND hydrosol and the supernatants after the nucleophilic addition reaction and washing of the composite. In preliminary experiments, we obtained absorption spectra of MND hydrosols with known concentrations of nanoparticles (Figure 4a). As for all MND hydrosols the optical density increased monotonically from the long wavelength range to the short wavelength range, we used the wavelength of 400 nm to plot optical density versus nanoparticle concentrations. The optical density of MND hydrosols linearly increased in a wide range of nanoparticle concentrations (0.025-1 mg/ml) (Figure 4b). Hence, this plot was used as a calibration curve to quantify MNDs in supernatants.
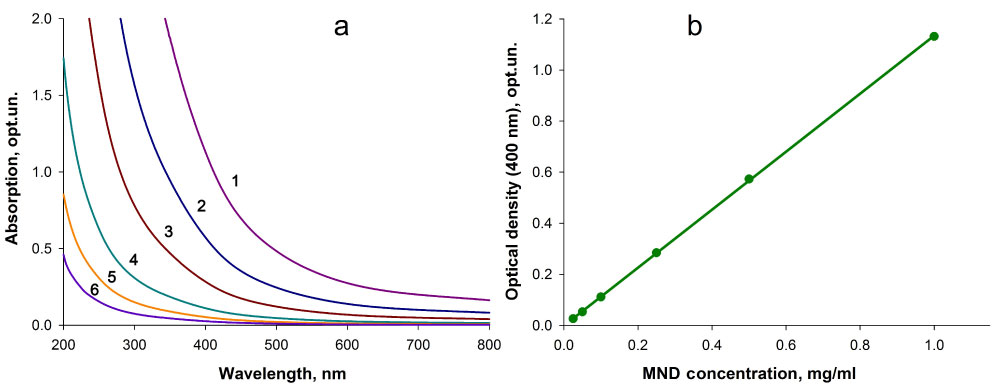 Figure 4: a) Absorption spectra of aqueous MND suspensions with different concentrations of nanoparticles (1-1 mg/ml; 2-0.5 mg/ml; 3-0.25 mg/ml; 4-0.1 mg/ml; 5-0.05 mg/ml; 6-0.025 mg/ml) and b) Optical density of MND hydrosols at a wavelength of 400 nm versus nanoparticle concentrations. View Figure 4
Figure 4: a) Absorption spectra of aqueous MND suspensions with different concentrations of nanoparticles (1-1 mg/ml; 2-0.5 mg/ml; 3-0.25 mg/ml; 4-0.1 mg/ml; 5-0.05 mg/ml; 6-0.025 mg/ml) and b) Optical density of MND hydrosols at a wavelength of 400 nm versus nanoparticle concentrations. View Figure 4
Calculations based on the data of spectral analysis of the initial MND hydrosol and supernatants after composite production and washing showed that under the experimental conditions (component ratio, suspension agitation speed, and time and temperature of incubation), up to 110 µg of nanoparticles can be covalently immobilized onto 1 mg of the polymeric carrier using the nucleophilic addition reaction.
In all cases studied, the adsorbed and covalently immobilized enzyme exhibited functional activity and catalyzed the formation of the colored product following the enzymatic and the non-enzymatic stages of assay were carried out. In the control experiments without the analyte or when the DEAE cellulose-MND composite material was used, the colored product was not observed.
Calculations based on results of spectral analysis of the initial urease solution and supernatants after covalent immobilization suggested that at the experimental conditions (pH and molar values of buffer solution, amount of DEAE cellulose-MND composite granules, temperature and duration of incubation), 1 mg of MND in the composite material bound 70-80 µg of the enzyme at the creation of urea determination system.
After covalent immobilization of urease onto the composite material, we conducted microscopic examination of the DEAE cellulose-MNDs-urease supramolecular system (Figure 5). Microscopy data analysis suggested that MNDs were rather uniformly distributed over the surface of the polymeric granules, giving them a brownish hue (Figure 5a). The microscopy of the supramolecular system in the fluorescence mode (after the enzyme had been labeled with the fluorescent dye) showed (Figure 5b) that being immobilized onto the DEAE cellulose-MND composite, urease was effectively bound to the surface of the carrier. Moreover, the microscopic image in the fluorescence mode demonstrated that the immobilized enzyme was firmly bound to the carrier without being desorbed during repeated washing of the supramolecular system.
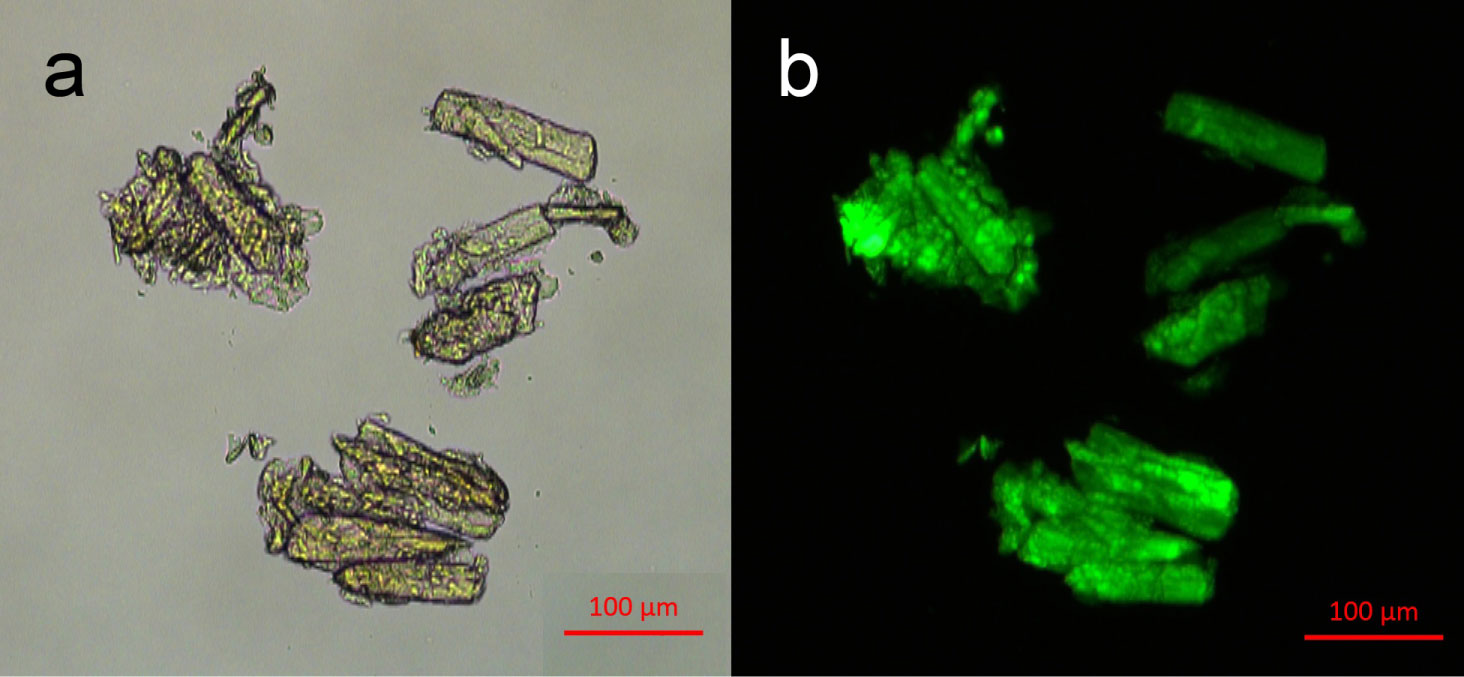 Figure 5: a) The appearance of the DEAE cellulose-MNDs-urease supramolecular system and b) Fluorescence of the FITC-labeled enzyme as a component of the supramolecular system. View Figure 5
Figure 5: a) The appearance of the DEAE cellulose-MNDs-urease supramolecular system and b) Fluorescence of the FITC-labeled enzyme as a component of the supramolecular system. View Figure 5
Experiments showed that urease molecules immobilized onto pure DEAE cellulose granules via adsorption or covalent conjugations were functionally active. However, when these indicator systems were repeatedly used to detect the analyte, the immobilized enzyme was considerably inactivated even at a temperature of 20-22 ºC, and that was evidenced by a decrease in the reaction product yield (Figure 6). A possible reason for this may be degradation of some of the enzyme molecules when the indicator systems were repeatedly settled by centrifugation and resuspended during numerous washing events, when residual substrate and reaction product were removed. At the same time, the enzyme covalently bound onto the DEAE cellulose-MND composite showed higher functional efficacy and enabled more stable yields of the colored product in repeated urea assays (Figure 6). It is worth noting that achieved in this work stable dynamics of the signals of DEAE cellulose-MND-urease indicator system at the multiple urea detecting is an advantage of the obtained system in comparison with similar urease sensors developed earlier. Urease immobilized on the DEAE cellulose - MNA composite lost only 8% its activity after six cycles of using the urea detection system (Figure 6). At the same time, in the other works urease immobilized on a grafted alginate beads, phosphonate grafted iron oxide nanoparticles, and calcium alginate beads lost about 17, 32, and more than 40% activity, respectively, after six cycles [38-40]. This allows us to conclude that the enzyme covalently immobilized on the MND particles of the composite is likely more resistant to damaging factors.
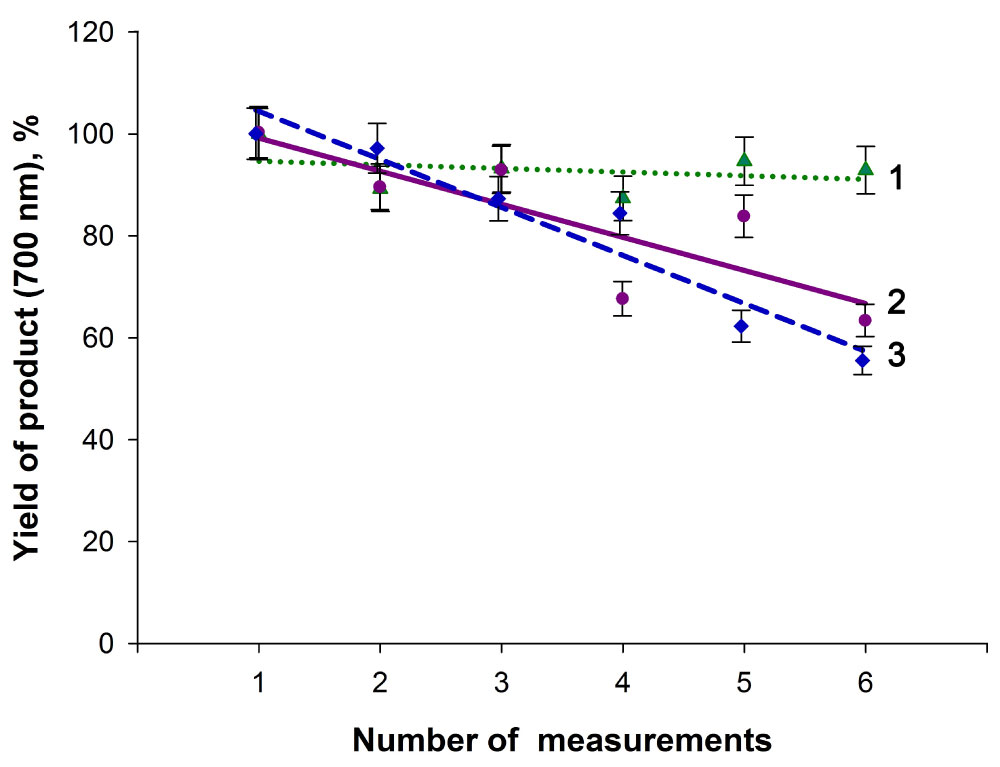 Figure 6: The yield of the reaction product in experiments with the supramolecular indicator systems repeatedly used to detect the analyte at 20-22 ºC. 1- The DEAE cellulose-MNDs- covalently immobilized urease supramolecular system, 2- The DEAE cellulose-covalently immobilized urease system, 3- The DEAE cellulose-adsorbed urease system. The data are normalized to the maximum yields of the product in measurement series. View Figure 6
Figure 6: The yield of the reaction product in experiments with the supramolecular indicator systems repeatedly used to detect the analyte at 20-22 ºC. 1- The DEAE cellulose-MNDs- covalently immobilized urease supramolecular system, 2- The DEAE cellulose-covalently immobilized urease system, 3- The DEAE cellulose-adsorbed urease system. The data are normalized to the maximum yields of the product in measurement series. View Figure 6
This assumption was supported by results of comparative experiments with the cellulose-urease and cellulose-nanodiamond-urease systems repeatedly used to detect urea at a temperature of 37 ºC (Figure 7). The enzyme covalently conjugated onto the DEAE cellulose-MND composite showed significantly greater stability towards the temperature factor, and its functional activity was reduced at a much slower rate than the activity of the enzyme covalently bound to the polymeric granules, as evidenced by the yields of the colored reaction product in repeated analyte assays (Figure 7).
 Figure 7: The yield of the reaction product in experiments with the indicator systems prepared by covalent immobilization of the enzyme repeatedly used to detect the analyte at 37 ºC. 1-The DEAE cellulose-MNDs-covalently immobilized urease supramolecular system, 2-The DEAE cellulose-covalently immobilized urease system. The data are normalized to the maximum yields of the product in measurement series. View Figure 7
Figure 7: The yield of the reaction product in experiments with the indicator systems prepared by covalent immobilization of the enzyme repeatedly used to detect the analyte at 37 ºC. 1-The DEAE cellulose-MNDs-covalently immobilized urease supramolecular system, 2-The DEAE cellulose-covalently immobilized urease system. The data are normalized to the maximum yields of the product in measurement series. View Figure 7
In the present study, the assay system for biochemical detection of urea was constructed to demonstrate the feasibility of constructing polymer-nanodiamond-biomarker supramolecular indicator systems. A composite support for immobilizing the enzyme was constructed from cellulose and MNDs. The comparative experiments showed that the supramolecular system produced by covalent immobilization of urease onto the DEAE cellulose-MND composite had the highest efficacy and enabled the most stable yields of the colored product in repeated analyte assays. The enzyme covalently bound to MNDs in the composite showed higher resistance to damaging factors and greater thermostability. The data obtained in the present study offer the prospect of designing a new type of reusable indicator assay systems (polymer carrier-nanodiamond-biomarker supramolecular systems) for biological and medical analytical applications. Further research is needed to optimize the conditions of constructing supramolecular indicator systems (including the systems on a two-dimensional polymer matrix and 3D polymer constructs) in order to enhance their efficacy and adapt the assay systems for practical use.