Uveitis is defined as inflammation of the uvea, which includes the iris, ciliary body, choroid, retina, and associated blood vessels. Non-infectious conditions, such as Vogt-Koyanagi-Harada illness and Bechet's disease, frequently cause uveitis. However, the pathophysiology of uveitis is also idiopathic. Unbalanced immune systems, genetics, and epigenetics all have a pivotal role in the development of disease. Several racial and ethnic groups strongly link HLA to BD (HLA-B51) and VKH (Vogt-Koyanagi-Harada) (HLA-DR4, DRB/DQA1). A study into the pathophysiology of uveitis claims a link between SUMO4, MCP-1, and CTLA4. A genome-wide association study (GWAS) showed that the genes IL23R/C1orf141, STAT4, and ADO/ZNF365/EGR2 all play a part in the development of uveitis. IL17F, IL23A, and C4A are examples of copy number variations (CNV) linked to uveitis. Additionally, epigenetic factors like DNA methylation and ncRNAs have a big impact on how the disease manifests itself. Exome, genome, and other epigenetic alterations may assist in identifying novel uveitis pathogenesis risk genes. Therefore, a deeper understanding of the condition's genetics could potentially improve uveitis management.
Uveitis, Vogt-Koyanagi-Harada, Behcet’s disease, Genetic predisposition, Single Nucleotide Polymorphisms (SNPs)
Uveitis is defined as inflammation of the uvea, which includes the iris, ciliary body, choroid, retina, and associated blood vessels [1]. We classify this condition as an intraocular inflammatory illness [2]. In uveitis, Behcet’s disease (BD) and Vogt-Koyanagi-Harada disease (VKH) are common non-infectious causes [3]. Both BD Behcet's disease (BD) and Vogt-Koyanagi-Harada disease (VKH) are autoinflammatory diseases. BD has recurrent oral aphthae, skin lesions, genital ulcers, and non-granulomatous uveitis. On the other hand, VKH has poliosis, vitiligo, alopecia, hearing problems, and granulomatous panuveitis on both sides and CNS abnormality [4-6]. The pathogenesis of uveitis is undetermined. People with more melanin, such as Asians and Native Americans, are prone to VKH disease, while BD, also known as Silk Road Disease, is prevalent in China, Japan, Korea, and Turkey [7,8]. A previous study demonstrates that human leukocyte antigens (HLA) in various racial groups closely correlate with the transmission of VKH sickness and Bechet's disease through families [9]. Researchers also found extreme variation in the clinical symptoms and onset ages of monozygotic twins with uveitis [10]. Integrating the data shows that epigenetic changes in association with an aberrant immune response are the primary causes of complicated genetic conditions or uveitis. Considering the fact that BD and VKH are both immune-mediated illnesses, the initial study primarily investigated the correlation with HLA polymorphism. HLA is largely located on the short arm of Chromosome 6p21.3 which has a length of around 4 × 10 -6 bp and contains several genes encoding proteins that also play a crucial and essential role in HLA. HLA can be categorised into three classes: class I, class II, and class III genes, based on the structure, tissue distribution, and functional variations of its coding molecules [11]. HLA-B51 is known to be a significant risk factor for BD across different ethnicities [12]. Researchers have found a significant link between HLA-DR4, DRB1/DQA1, and VKH disease among various ethnic groups [13]. Despite the substantial link between HLA, Bechet's illness and VKH disease, researchers estimated that HLA only accounts for 19% of the total genetic susceptibility to both diseases. Many studies then looked at the correlation between the above-described parameters and non-HLA genes. With VKH sickness and Behcet's disease. This review covers the most recent findings about the relationship between non-HLA genetic or epigenetic variables and the diseases BD and VKH.
Single-site variations, like single nucleotide polymorphisms (SNPs), and structural variants, like copy number variations (CNVs), are two categories of genetic variants. Variants vary from minor base changes to significant fragment mutations [14].
SNPs make up less than 1% of the human genome, while big DNA sequence variations, also known as copy number variations, make up between 5 and 10% of the genome. These variations encode different copies of a particular gene. These CNVs can account for around 10-20% of the heritability of specific diseases since their mutation rate is substantially higher than that of SNPs.
Although the cause of their aberrant expression is unclear, other investigations have indicated that patients with uveitis express a variety of inflammatory factors dysfunctionally. It was found that there is link between copy number variations (CNVs) in IL17F, IL23A, and C4A and BD and VKH disease to learn more about their genetic basis. Outcomes are depicted in Figure. Refer to Figure 1 [15].
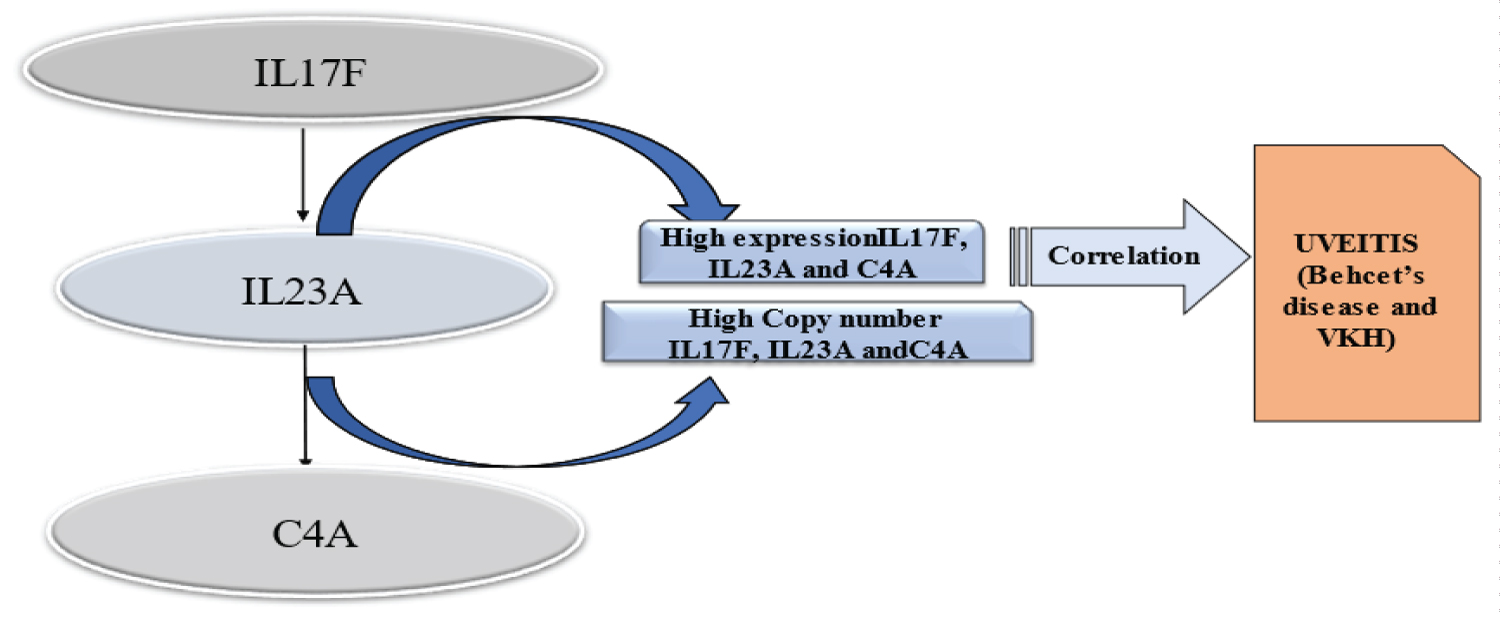 Figure 1: This picture shows the results, which showed a link between higher levels of IL17F, IL23A, and C4A expression and high copy numbers of IL17F, IL23A, and C4A in people with Bechet's illness or VKH disease. This suggests that these CNVs play a part in the development of uveitis.
View Figure 1
Figure 1: This picture shows the results, which showed a link between higher levels of IL17F, IL23A, and C4A expression and high copy numbers of IL17F, IL23A, and C4A in people with Bechet's illness or VKH disease. This suggests that these CNVs play a part in the development of uveitis.
View Figure 1
Single nucleotide polymorphisms or SNPs are the most common and abundant form of genetic variation in humans. It was evaluated more than 3 million SNPs in the human genome. Sequence variations of the genome spawned by single nucleotide base pair alternatives generate sequence variations in the genome. 90% of phenotypic variations are due to SNPs. The human chromosome 2q33 of the CD28 gene family has a cytotoxic T-lymphocyte-associated antigen (CTLA-4). The latest studies demonstrate the relationship between CTLA4 gene polymorphisms (-1661A/G, -318C/T, +49G/A, and CT60) with BD and VKH diseases in the Chinese Han population [16,17]. VKH aligned with haplotypes -1661A, -318C, +49G, CT60G and +49G align with VKH but did not align with BD. Therefore, VKH and BD show genetic heterogeneity. Clinical presentations of both diseases vary; BD is non-granulomatous inflammation, whereas VKH is granulomatous inflammation [18]. The preceding data depicts the association between CTLA4 gene polymorphism and BD in different ethnic cohorts. Small ubiquitin-like modifier 4 (SUMO4) is found on chromosome 6p25. SUMO4 controls the transcription of nuclear factor kappa B (NF-kB). SUMO4 is involved in a plethora of autoimmune diseases, such as type 1 diabetes [19]. Vascular-related diseases like diabetic neuropathy and diabetes mellitus associated uveitis (DMAU) typically complicate diabetes [20]. In the Chinese Han population, association of the SUMO4 +438 C allele and AGAT haplotype with BD was ruled out [21]. Investigations performed by Korean and Tunisian groups substantiate the analysis [22]. SUMO 4 happens to be an unsafe gene for vascular autoimmune diseases like BD and Type 1 diabetes. A genome-wide association study (GWAS) is a fair and dynamic tool for determining any genetic variant associated with human diseases. A GWAS was conducted on Bechet's patients in Turkey. Five related genes was ruled out, including KIAA1529, CPVL, LOC100129342, UBASH3B, and UBAC2. Further investigations revealed that UBAC2, among the five mentioned genes, was associated with BD in, Italian, and Japanese populations. Therefore, UBAC2 may be a common hazardous gene for various ethnic populations. Furthermore, researchers examined genetic predispositions related to VKH and BD in Chinese Han individuals. It was observed a close relationship between BD and STAT4. The presence of SNP rs897200 in STAT4 affects the binding of STAT4 with transcription factors like YY1 and POUIF1a, as demonstrated by bioinformatics and functional studies. It was observed an intensification of STAT4's transcriptional potential, which further triggers the release of proinflammatory factors like IL-17, clearly demonstrating STAT4's involvement in BD through Th17 pathway regulation. Researchers found a significant association between BD and STAT4 in Korean and Turkish groups [23]. GWAS analysis perforate large sample population's GWAS analysis revealed VKH's association with numerous non-HLA genes, including IL23R/C1orf141 and ADO/ZNF365/EGR2 study was conducted in Singaporean, Thai, and Korean populations to rule out the relationship between VKH and the IL23R/C1orf141, EGR2/ADO, and ZNF365 genes. IL23R/C1orf141 is associated with Singaporean VKH patients [13], whereas EGR2/ZNF365/ADO is associated with VKH in the Thai population. An intriguing aspect of the study was the association of EGR2/ADO with BD in Chinese Han and Turkish ethnic groups [24].
'Epigenetics' is the term for the branch of genetics that does not include DNA sequencing. It focuses on factors such as DNA methylation, histone modification, and non-coding RNA (ncRNA) that may alter gene expression. The process of DNA methylation involves the precise covalent attachment of methyl groups to DNA bases and the cytosine of CpG dinucleotides. In the case of Bechet's, recent studies explain DNA methylation in monocytes and CD4+ T cells. Researchers evaluated 383 differentially methylated CpG sites in monocytes and 125 differentially methylated CpG sites in CD4+ T cells in patients with BD [25]. Another DNA methylation study suggests a link between 4332 differentially methylated CpG and BD. A replication study suggests an association between BD at 5’UTR and FKBP5 [26]. There is a high prevalence of GATA3, IL-4, and TGF-beta in patients with VKH disease [27]. Therefore, DNA methylation plays a role in the development of both BD and VKH. Types of ncRNA are ncRNA, miRNA, and circRNA, classified based on their sequence length. BD results in a decrease in miR-155 expression, while VKH strongly associates with variations in the copy number of miR-23a, miR-146a, and miR-301a [28]. The overexpression of miR-23a in ARPE-19 cells activates proinflammatory cytokines IL-6. Thus, research has proven that ncRNAs play a vital role in a normal immune response that leads to BD and VKH [29].
A variety of clinical symptoms, commonly including extraocular and ocular sites, characterize uveitis, a complex multisystem disease. This study examines VKH and BD, the two most significant genetic risk factors for uveitis. Although uveitis has an idiopathic etiopathogenesis, Evidence suggests that a combination of certain genetic or epigenetic factors may be responsible for an imbalance in immune response regulation, leading to the development of uveitis. Researchers have drawn an association between VKH and BD with specific HLA types, numerous SNPs or CNVs in non-HLA genes, ncRNAs, and DNA methylation in various ethnic communities. A tiny fraction of identified genes accounts for genetic uveitis, and we will soon discover many more genes responsible for the condition. The application of various modern technologies, such as whole genome sequencing and whole exome sequencing, as well as the analysis of certain other epigenetic factors, such as N6-methyl-adenosine modification of mRNAs, will be helpful in the identification of new pathogenic risk genes for uveitis. Understanding the genetic and epigenetic mechanisms of uveitis may assist in developing a foundation for the discovery of new targets and may lead to the development of novel strategies in the treatment of uveitis in the coming future.
Not applicable.
Authors have no conflicts of interest to declare.
Not applicable.
Not applicable.
Not applicable.
I extend my heartiest gratitude towards my parents, a special thanks to our guide Madhvi Ghadhge Ma’am, Vartika Jain, Pallavi and all other co-authors for immense support throughout the article.
SB(Sagnika Bhattacharjee), SG(Sakshi Gupta), SC(Supta Chakraborty), MG(Madhvi Ghadhge), Pallavi, Dr VJ (Vartika. Jain) have equally contributed to the article, SB(Sagnika Bhattacharjee) analyzed previous article, reviewed them, prepared the discussion and conclusion section and contributed in overall refining of the manuscript, SC(Supta Chakraborty) has prepared the images, SG(Sakshi Gupta has written some portion of manuscript ), Dr. Vartika Jain (VJ), Madhvi Ghadhge (MG), Pallavi has assisted and guided in preparing the manuscript.
All the authors have equally contributed. The authors have read and approved the final manuscript.