Brain imaging quality of data is important to confirm their liability of brain imaging studies. Previous literature confirmed that some confounding factors such as movement, age, and gender may impact brain imaging quality. Automatic quality control (QC) applications may not be able to properly calculate their reliability due to confounding factors.There are a few studies on brain imaging quality data and relevant confounding factors such as age or gender.
Open data from a previous study was used to conduct this study. In total 26 participants were recruited. Random Forest (RF) and Neural Networks (NN) machine learning (ML) methods were used to predict age groups (cut-off age: 16). Patients were grouped by age groups. Then, the age group was predicted with RF and NN machine learning (ML) models.
The goal of the study was to predict age groups using brain imaging quality data.
We found that according to NNs, the age group was predicted with an accuracy of over 60% (accuracy: 64%, sensitivity: 50%, specificity: 71%, area under curve (AUC): 55%,). Furthermore, the RFML model found that the age group was predicted with an accuracy of 64% (sensitivity: 50%, specificity: 71%, AUC: 86.6%).
Our study showed that age groups can be predicted using the brain imaging quality of the data. Further studies should investigate the relationship between other brain imaging parameters related to the quality of data and age.
Schizophrenia, Brain imaging, Brain imaging quality, Prediction, Age, Brain age, Visual inspection
SCZ: Schizophrenia; NN: Neural Networks; RF: Random Forest; QC: Quality Control; MRI: Magnetic Resonance Imaging; HC: Healthy Controls; fMRI: Functional MRI; ML: Machine Learning
Neuroimages must be checked for potential distortion after processing [1]. Previously, the impact of data quality on the localization of brain activation in functional magnetic resonance imaging (fMRI) was investigated in-depth [2]. Also, some studies concluded that low-quality imaging data may cause false associations. Many techniques such as deep learning [3] and random forest were used to calculate the image quality of the human brain data whilst age is related to in-scanner motion and data quality [4]. Furthermore, several factors are related to MR image quality assessments such as geometric accuracy, high-contrast resolution, slice thickness accuracy, and slice position accuracy [5]. It is also important to analyze the association of parameters with age groups since it is one of the potential confounding factors.
Taken together, this study aimed to predict age groups using brain imaging quality data created using MRI QC.
To provide measures describing the quality of this dataset, the data were analyzed using MRIQC [3]. The open fMRI Data from the previous study was used to conduct this study.
This data was obtained from the Open fMRI database. The fMRI data is available at (https://exhibits.stanford.edu/data/catalog/xg798vw8719) [6].
MRIQC was used to measure the quality of brain imaging.
In this study, Neural Networks (NN) and Random Forest (RF) were used to predict age groups.
Neural networks (NN) predicted age groups. Scikit-learn default parameters were used to create prediction models [7].
Random forest: Random Forest (RF) [8] is one of the popular ML algorithms used to predict age groups in this study. Scikit-learn default parameters were used to create prediction models [7].
Participants were selected from the Open fMRI data data base. There was no missing value to report based on the shared data. The threshold of 0.05 was considered significant. Independent Sample T-test and Mann Whitney tests were used according to the normality of the study. Scikit-learn package of Python was used to create ML model [7].
In this study, the following brain imaging quality parameters were used to predict age groups: summary_mean_bg, summary_mean_csf, summary_mean_gm, summary_mean_wm, summary_p05_bg, summary_p05_csf, summary_p05_gm.
summary_p05_wm, summary_p95_bg, summary_p95_csf, summary_p95_gm, summary_p95_wm, summary_stdv_bg, summary_stdv_csf, summary_stdv_g, and summary_stdv_wm.
The mean age of the all participants was "16.89''. The standard deviation was ''4.26''. The minimum age was ''8.8'' while the maximum age was ''25.6''. Most of the patients who participated in this study were female (n = 14) while 13 of the patients were male.
In this study, random forest (RF) and neural networks (NN) were implemented to predict age groups.
Random forest: The RF ML model found that the age group was predicted with an accuracy of 64% (sensitivity: 50%, specificity: 71%, AUC:86.6%). The most important predictive features were Summary_p05_wm, summary_p05_csf, summary_stdv_csf, summary_mean_wm_, and summary_p05_gm respectively. For the RF model mean cross-val accuracy was 61 ± 0.12.
Neural networks: Based on the NN model, the accuracy of ML model was 64% (sensitivity: 50%, specificity: 71%, AUC: 55%). The most important predictive features were Summary_p95_csf, summary_mean_wm, summary_p95_bg, summary_mean_csf, and summary_stdv_bg respectively. For the NN model mean cross-val accuracy was 58.6 ± 0.26.
Based on our main results of the study, we found that age groups can be predicted with both Random Forest (RF) and Neural Networks (NN) with the same accuracy over chance level.
Brain imaging data is not usable for extracting meaningful information without processing [9] and it should have some quality standards to be used.
The performance of the quality control (QC) strategies depended on the morphological measure [10]. Automated QC is important where visual QC is not practical [11]. Manual quality control strategies are reliable for QC of brain segmentation [10] however, manually controlling may take time. Furthermore, QC protocols allow different laboratories to search the impact of QC on the relationship between the brain and phenotypes [12].
Besides, The degree of anonymization of the data is important [13]. Revealing brain imaging quality data may allow researchers to predict their age groups. Furthermore, the age-dependent increase was found in brain because of the changes related to myelination.
Consistent with the findings that activity from almost all areas of the brain became less predictable with increased age [14]. Moreover, the ability to evaluate MRIs for disease characterization is hampered by the artifacts [15].
Studies on such "brain age prediction" vary widely by their methods [16]. Developmental processes that occur during the first two decades of human development impact brain development (e.g. body growth and puberty) [17]. In addition, Age can be a confounding factor in some studies [18]. Further, the percent signal change associated with the BOLD effect increases with age in children ages [19]. Taken together, age is one of the important confounding factors that potentially impact brain imaging quality data.
This study concluded that age groups can be predicted with high accuracy with RF and NN ML algorithms using brain imaging quality data.
Further studies should investigate other confounding factors that may affect brain imaging quality.
There were several limitations in the current study. Gender was one of the important confounding factors.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Thanks to the participants who made this study possible.
There is no funding associated with this study. KU was supported by Chinese Government Scholarship provided for PhD students.
The MRIQC Web API is available under the Apache-2.0 license. The source code is accessible through GitHub (https://github.com/poldracklab/mriqcwebapi).
The fMRI data is available at (https://exhibits.stanford.edu/data/catalog/xg798vw8719) [6] (Table 1, Table 2, Table 3, Table 4) and (Figure 1, Figure 2).
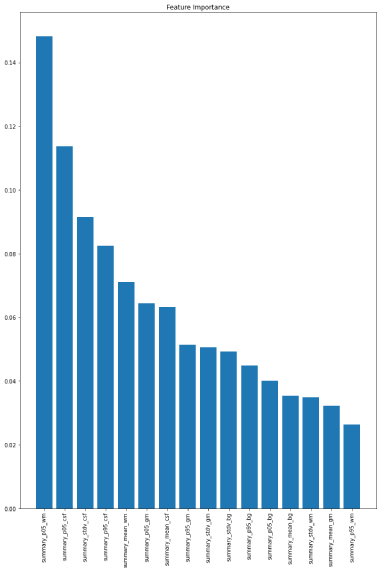 Figure 1: Feature importance's associated with random forest model.
View Figure 1
Figure 1: Feature importance's associated with random forest model.
View Figure 1
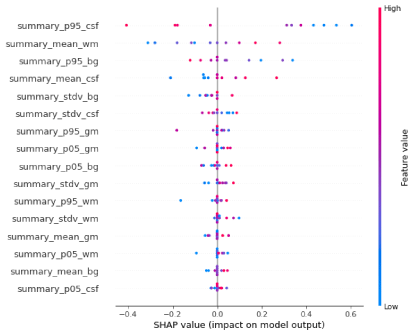 Figure 2: Feature importance's associated with neural networks model.
View Figure 2
Figure 2: Feature importance's associated with neural networks model.
View Figure 2
Table 1: Confusion matrix associated with random forest model. View Table 1
Table 2: Confusion matrix associated with neural networks model. View Table 2
Table 3: Accuracy scores (Random forest). View Table 3
Table 4: Accuracy scores (Neural networks). View Table 4
Kadir Uludag is funded by Chinese Government Scholarship Type-A.
No potential competing interest was reported by the authors.