Trypanosomosis is a major neglected disease of animals and man that causes great negative socio-economic impact in many African countries. It is caused by protozoan parasites of the blood from the genus Trypanosoma. Previous studies have investigated the prevalence and risk factors of trypanosomosis in Tanzania, but none has been done in the human/livestock/wildlife interface areas of Mikumi National Park. The present study determined prevalence of trypanosomosis in cattle blood sampled in five villages of the Mikumi interface areas. Trypanosome species were identified using the nested ITS-PCR and SRA-LAMP. Acquired biodata and spatial information were used for the analysis of risk factors based on the proximity from the park. Data analysis for categorical variables was performed using Chi-square, Fishers exact test, Odds and Risk ratios. Maps were created using the ArcMap™ version 10.1 GIS software (ESRI 2012). Overall prevalence was 51.47%. Infection significantly varied among the 5 villages (p = 0.0022). The study identified 7 different species of trypanosomes. Trypanosoma simiae had the highest rate of infection 48.15%, followed by T. theileri 26.67%, T. vivax 16.30%, T. brucei brucei 4.44%, T. congolense forest 2.22%, T. simiae tsavo 1.48% and T. congolense kilifi 0.74%. No zoonotic species were identified. High density vegetation (p = 0.0001) and human activity (livestock market and cattle movement) (p = 0.0001) were identified as strong risk factors for infection. Areas with charco dams (p = 0.0001) and those close to tsetse screens and traps (p = 0.0006) had significantly lower infection prevalence. Identifying risk factors, quantifying the risk and doing spatial analysis is essential to determine the control measures to be used in the affected rural communities.
Trypanosoma spp, Infection, Interface, Risk, Zoonotic
Trypanosomosis is a major neglected vector borne disease of animals and man with a great negative socio-economic impact [1-3]. It is present in more than 37 African countries [2]. Caused by protozoan parasites from the genus Trypanosoma that inhabit the blood stream, lymph nodes and other tissues [4,5]. Different species of trypanosomes affect a wide variety of animal species including mammals, birds, reptiles and fish [6,7]. Trypanosoma congolense, T. brucei, T. vivax, T. simiae, T. godfreyi and T. theileri are some of the species circulating in domestic and wild mammals of Sub-Saharan Africa [8-10]. T. congolense, T. brucei and T. vivax have been identified in a wide range of wild bovids and suids, T. congolense has also been reported in carnivores (Panthera leo and Crocuta crocuta), T. theilery in wild bovids while T. simiae and T. godfrei have been identified in suids (Phacochoerus africanus) [8,9]. Animal African Trypanosomosis (AAT) or nagana in cattle is commonly caused by T. congolense, T. vivax and T. brucei [10]. Human African Trypanosomosis (HAT) caused by T. brucei gambiense and T. brucei rhodesiense, is also a re-emerging neglected disease [10-12]. Although the animal pathogens do not infect humans, domestic and wild animals can serve as reservoirs for zoonotic trypanosomes [13].
The epidemiology of trypanosomosis is strongly related to the ecology of the tsetse fly [6]. Although some trypanosome species can be transmitted in other ways, the tsetse fly is the main cyclic vector of trypanosomosis [14,15]. The tsetse are insects from the genera Glossina [5], found in sub-Saharan Africa between latitudes 14 °N and 29 °S, occupying more than 10 million km2 in specific environments defined by climate, vegetation and fauna [7,16].
The existence of wildlife reservoirs and vectors increases the risk of infection in communities surrounding parks and reserves [13,17]. Climatic and ecological changes [7,18], pastoralism and rapidly increasing human population invading wilderness areas [10,19] also influence infection rates.
Mikumi National Park is the fifth largest park in Tanzania (3230 km2) and interactions commonly occur in and around it [20]. Nevertheless there is a gap in knowledge regarding the status of trypanosomosis in its human/livestock/wildlife interface areas, which encouraged a wider, collaborative and multidisciplinary (one health) approach to the issue [21].
The main objective of this study was to identify the trypanosome species circulating in pastoral cattle in the human/livestock/wildlife interface areas of Mikumi National Park. Specifically this work aimed at (i) determining the spatial prevalence of trypanosome species in cattle grazed in the Mikumi interface areas, as well as (ii) screening for presence of human-infective trypanosomes in cattle and lastly (iii) assessing risk factors for human and animal infection.
In this cross sectional study a total of 237 samples were collected from 74 bomas in five selected villages (Mikumi, Ihombwe, Kidui, Mbwade and Parakuyo) in the interface areas of Mikumi National Park, Kilosa District, Morogoro Region [20,22-24]. Random stratified sampling was carried out considering the total number of cattle of each village. Sample size was determined [25] using a 1.25 design effect, 2.3% prevalence previously reported for Morogoro [26], 6% absolute error and 95% Confidence Interval (CI). Animals above one year of age were randomly selected for the study and categorized as immature (1-3 years) and adult (> 3 years). Biodata was registered in biodata sheets during the sampling process. The sampling was carried out in July 2016, which was in the dry season. Blood samples (5 ml) were collected from the jugular vein into EDTA vacutainer tubes and stored in refrigerated conditions (4 °C) until arrival at the laboratory, where they were stored at -20 °C until processing time.
Genomic DNA was extracted from whole blood using the Zymo Research protocol as described in Quick-g DNA™ Blood miniprep (D 3073) and stored at -20 °C until further analysis.
Nested ITS-PCR was conducted using the ProFlex™ PCR System Thermocycler (Applied Biosystems®, Life technologies) in order to detect infection and allow the identification of trypanosome species according to band size (Table 1) [27].
Table 1: Cycling conditions for nested ITS-PCR and expected band sizes for each Trypanosoma species. View Table 1
Eluate (2.5 μL) containing extracted DNA was added to 12.5 μL of PCR master mix with the outer ITS primers ITS1 (5' - GAT TAC GTC CCT GCC ATT TG - 3') and ITS2 (5' - TTG TTC GCT ATC GGT CTT CC - 3') (Inqaba Biotec). The second round of PCR which contained the inner ITS primers ITS3 (5' - GGA AGC AAA AGT CGT AAC AAG G - 3') and ITS4 (5' - TGT TTT CTT TTC CTC CGC TG - 3') (Inqaba Biotec) was seeded with PCR products (0.5 μL) from the first round. A positive control of T. simiae was used. The negative control consisted of water instead of eluate [27,28].
For electrophoresis 1.5% (w/v) agarose gels were prepared in 1 x TAE buffer (0.04 M Tris-acetate, 0.001 M EDTA, pH 8.0) (AMRESCO®) and stained in 5 μL GR Green (Excellgen). The gels along with 100 bp molecular weight Quick-Load DNA Ladder (Quick-Load®, New England Biolabs, Inc.) (0.5 μL /lane) were run for 40 min at 80 volts and fluorescent gel images were visualized using the E-Box imaging system (E-Box C X 5, Vilber Lourmat). When the amplification reactions showed expected band sizes and no reaction for the negative control, infection was considered present.
SRA-LAMP technique was used in all T. brucei positive samples in order to detect human infective T. b. rhodesiense. A 25 μL reaction volume was prepared to conduct the SRA-LAMP. The reaction mix contained 2.5 μL of 1 x Thermopol buffer, 5 μL of 0.8 M Betaine, 1 μL of 0.2 μM SRA-F3 (5' - GCG GAA GCA AGA ATG ACC - 3') and SRA-B3 (5' - TCT TAC CTT GTG ACG CCTG - 3') primers, 1 μL of 2 μM SRA-FIP (5' - GGA CTG CGT TGA GTA CGC ATC CGC AAG CAC AGA CCA CAG C - 3') and SRA-BIP (5' - CGC TCT TAC AAG TCT TGC GCC CTT CTG AGA TGT GCC CAC TG - 3') primers, 1 μL of 0.8 μM loop primers SRA-LB (5' - GCA GCG ACC AAC GGA GCC - 3') and SRA-LF (5' - CGC GGC ATA AAG CGC TGAG - 3'), 1 μL of 8 U Bst DNA polymerase, 1 μL of 200 μM dNTPs, 7.5 μL of nuclease free water and 2 μL of DNA template [29]. SRA-LAMP ran in a GeneAmp® PCR system 9700 (Applied Biosystems™, USA). Reaction conditions were set at 62 oC for 1 hr and terminated by raising the temperature up to 80 oC for 5 mins.
Two microlitres (2 μL) SYBR® Green I were added to the SRA-LAMP products promoting color visualization. Color alterations to green signalled a positive reaction and color change to orange indicated negative results. Results obtained were confirmed by gel electrophoresis where 10 μL of product was run on 1.5% (w/v) agarose gel at 100 volts for 30 mins.
Epi Info™ version 7.1.4.0 (CDC, Atlanta, USA) and Microsoft Office Excel 2007 were used to statistically analyze the data collected from each study animal. The prevalence of trypanosomosis was calculated for each village. The Chi-square test and Fishers exact test were used to analyze categorical variables. A p-value lesser than 0.05 (p < 0.05) was considered significant. Odds ratio and Risk ratio were used to determine the strength of association of risk factors with the occurrence of infection. Spatial descriptive maps of the trypanosome species distribution were produced using the ESRI 1999-2012, ArcMap™ version 10.1, GIS software (Environmental Systems Research Institute, USA).
Satellite and GIS generated figures using ArcMap™ version 10.1 GIS software ESRI 2012 revealed that Ihombwe, Kidui and Mikumi villages were located closer to the park, with distances of approximately 6 km, 4 km and 3 km from the park, respectively, while Mbwade and Parakuyo were located at a distance of approximately 20 km and 24 km from the park, respectively (Figure 1).
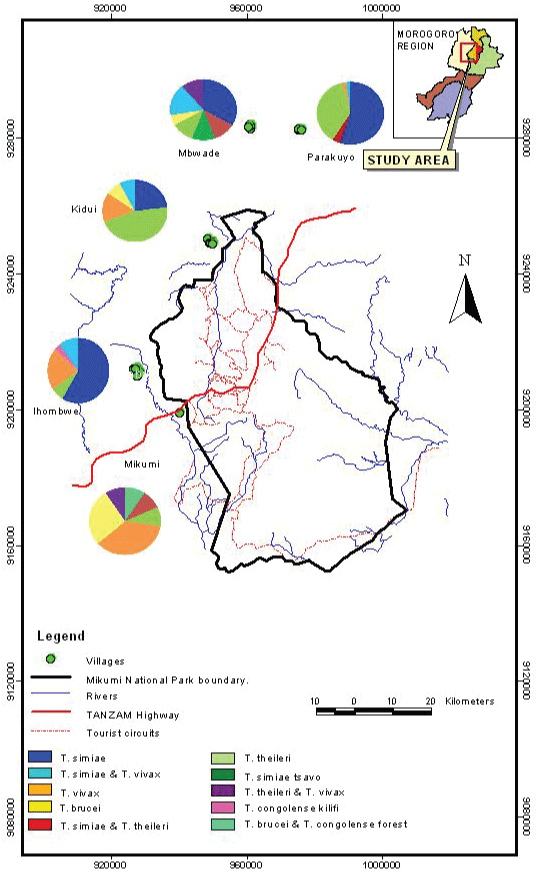 Figure 1: Map showing distribution of trypanosome species per village. View Figure 1
Figure 1: Map showing distribution of trypanosome species per village. View Figure 1
Further, it was revealed that extensive farming system is practiced in all villages allowing the interaction with cattle from other locations as well as wildlife. Two main ethnic groups were identified: Mbwade, Parakuyo and Kidui were predominantly inhabited by the Maasai, while Ihombwe was a typical Sukuma village and Mikumi had a mixture of both permanently residing Sukuma and temporary Maasai camps for keeping cattle in the dry season. During the dry season there is a scarcity of pastures and no supplements are given to the animals. Many Maasai livestock keepers migrate with their cattle to temporary grazing locations where they camp leaving a few animals behind at the house hold, especially lactating cows, their calves and old animals incapable of grazing far. Families also keep other animal species such as small ruminants and dogs.
Out of the 237 cattle examined, infection was detected in 122 animals, with an overall prevalence of 51.47%. All surveyed villages registered infection. Prevalence of infection significantly varied among villages, Ihombwe had the highest prevalence of 67.44%, followed by Mbwade 56.25%, Parakuyo 55.43%, Mikumi 50% and Kidui 27.08% (Χ2 = 16.7107, df = 4, p = 0.0022).
Out of the infected animals 86.07% presented a single trypanosome species in circulation and the remaining 13.93% had infection caused by two different trypanosome species simultaneously. The village with highest rate of single infections was Parakuyo (94.12%) and Mbwade had the highest rate of double infections (38.89%). Further details are shown in Table 2.
Table 2: Single and double infections per village. View Table 2
Seven species of trypanosomes were identified and the specific distribution of the parasites is detailed in Table 3. Overall Trypanosoma simiae had the highest rate of infection 48.15%, followed by T. theileri 26.67%, T. vivax 16.30%, T. brucei 4.44%, T. congolense forest 2.22%, T. simiae tsavo 1.48% and T. congolense kilifi 0.74%. Ihombwe, Mbwade and Parakuyo had a higher occurrence of T. simiae compared to all the other species. In Kidui T. theileri was the most predominant and T. vivax occurred at a higher proportion in Mikumi village.
Table 3: Prevalence of trypanosome species identified per village. View Table 3
This study revealed a 4.44% prevalence of T. brucei in 3 (Kidui, Mbwade and Mikumi) out of 5 villages. The highest rate of infection was registered in Mikumi village. In order to further characterize whether the PCR-identified T. brucei parasites were human infective the SRA-LAMP technique was employed. None of the 6 T. brucei positive samples were shown to be T. b. rhodesiense. All were T. brucei brucei.
Factors considered relevant for trypanosome infection in cattle are shown in Table 4.
Table 4: Analysis of the association of demographic and other factors with trypanosome infections in cattle. View Table 4
Occurrence of infection did not show statistically significant variation in cattle of different sexes, age groups, body condition or breed, as well as in cattle whose owners were pursuing different types of main activities and in cattle from different sources. Although odds and risk ratios showed that older animals, animals with poor body condition, Ankole and mixed breeds, cattle kept in homesteads where people engaged mainly in crop production and those transferred from other areas were more likely to harbour the parasite.
On the other hand infection was significantly higher in cattle from villages that had easy access to livestock markets (p = 0.0001) and the odds of occurrence and risk of infection were equally high (OR = 3.6683, CI = [1.8235-7.3792], RR = 1.7227, CI = [1.3556-2.1891]).
Results indicate that farmers treated their animals using one or more of combinations of trypanocidal drugs without professional supervision. Furthermore, use of the different drugs or their combinations (Berenil®, Berenil® and Oxytetracyclin, Samorin®, Novidium®) did not seem to significantly influence the infection occurrence. This study also revealed that farmers used two main groups of insecticides/pesticides for spraying cattle: pyrethroids (Cybadip®, Albadip®, Paranex®, Dominex®, Karatii®) and organophosphorus compounds (Steladone®). Infection was found in 51.95% of the cattle under pyrethroid treatment and 30% of cattle treated with organophosphorus compounds, although variations were not statistically significant within each group organophosphorus compounds displayed a protective effect.
An interesting finding was revealed when water sources used by cattle were investigated. Thus, cattle drinking from rivers and water wells were shown to be at higher risk of trypanosome infections compared to those drinking from charco dams (p = 0.0001). Vegetation cover was shown to have a significant role on infection rates in all villages. Significantly lower trypanosome infections (p = 0.0001) were found in Kidui where vegetation like the open miombo woodlands is fairly sparse compared to the other villages where vegetation was denser.
Proximity to Mikumi National Park did not show a significant variation of infection rates among the five villages although the villages closer to the Park had a slightly lower occurrence of infection (46.9%) when compared to the farthest villages (55.64%). Villages located closer to the tourist tracks which contain treated tsetse traps and screens showed significantly lower infection occurrence (p = 0.0006).
The present investigation reports an overall prevalence of 51.47% for trypanosome infections in cattle in Mikumi interface areas based on nested ITS-PCR. The high prevalence may be explained by the high sensitivity of nested ITS-PCR employed in this study as well as the ecology of the study area, where wildlife and livestock intermingle creating conditions for the circulation of the trypanosome parasites. This prevalence may be considered high when compared to findings in studies based on parasitological methods but resembles other molecular studies. A study carried out in Morogoro region [26], showed the prevalence to be of 2.3% by parasite microscopic identification methods. A different parasitological survey for bovine trypanosomosis based on microscopic examination done in Arusha Region, Northern Tanzania, detected infection levels ranging from 3% to 7% [30]. In Kenya, a prevalence of 28.1% was reported using nested ITS-PCR, 26.2% using single ITS-PCR and 10.7% by using the species specific PCR for the same samples [31]. A different study in Kenya using species specific primers showed a prevalence of 24.1% in cattle and 25.9% in camels [32]. A prevalence ranging from 6% to 57.1% for T. evansi was found in camels from different regions of Sudan using the ITS1 primer-based PCR [33]. In Uganda a comparative study done in dairy cattle, determined prevalence of 34.8% using species specific PCR, 9.3% using the haematocrit centrifuge technique and 16% via the mini-anion-exchange technique [34].
Several studies have reported prevalence and risk factors of Animal African Trypanosomosis (AAT) and Human African Trypanosomosis (HAT) in many African countries including Tanzania [35-39].
Clinical and conventional parasitological diagnostic techniques are commonly used to detect trypanosome infections in African countries due to their simplicity [30,40]. Nevertheless, recently molecular techniques have been gaining more appreciation by the scientific community [41].
Nested ITS-PCR was used in the present study due to its high sensitivity and specificity, rapid results and the ability to identify a wide range of trypanosome species, thus resulting in the diagnosis of species that usually are not diagnosed in routine surveys. A study comparing nested ITS-PCR, single ITS-PCR and species specific PCR as diagnostic assays for the detection of pathogenic trypanosomes in cattle blood showed higher sensitivity of the nested ITS-PCR comparing to the others techniques [31]. Another advantage of the ITS-PCR is the ability of multi-species detection of all clinically important species and some subspecies making it suitable for large-scale epidemiological studies [28,31].
The SRA-LAMP technique used to screen for human infective trypanosomes in all the positive T. brucei samples in this study is a novel molecular technique described as very sensitive, reproducible and user friendly [29]. On the other hand the use of species specific PCR is recommended as a more specific tool for identification of the trypanosoma members when comparing to ITS-PCR [27]. Different types of PCR have also been used to detect trypanosomes in tsetse flies [9,11,36,42-44] and in human blood [45].
In the present study 48.15% of all detected infections were due to T. simiae and 26.67% to T. theileri, greatly increasing the general prevalence. These findings are similar to another study that demonstrated high prevalence of T. theileri (29.86%) and T. simiae (1.38%) through nested ITS-PCR along with the common T. brucei, T. congolense and T. vivax [28]. The overall prevalence for the same samples increased from 28.47% using single ITS-PCR to 63.88% using nested ITS-PCR [28]. The nested ITS-PCR in the present study allowed the detection of a greater number of species, identified double infections and provided a better picture of infection distribution for comparison among studied villages.
High prevalence of T. simiae in our study area is probably due to the presence of wild suids and other wildlife reservoirs in the Mikumi interface [46,47]. Mechanical transmission and presence of Tabanids may also be a cause of high T. theileri and T. vivax counts in the area [48].
The high prevalence of infection found in some villages further from the park may indicate the perpetuation of an infection/re-infection cycle between domestic animals and wild animals, where both serve as reservoirs. The lower infection in some villages closer to the park may be due to the significantly protective effect (p = 0.0006) of being close to the tourist tracks where insecticide impregnated screens and traps are positioned every 100 m.
In a study in Morogoro, it was observed that domestic animals living and grazing closer to the interface areas were at higher risk of acquiring infection [26]. The report showed that male animals, animals with poor body score and animals above 4.5 years of age were at higher risk of infection [26]. The findings from the present study have further shown that distance from the park is not a main risk factor, instead type of vegetation cover and human behaviour and activity significantly increase the risk to infection. These risk factors most likely provide the vector with an appropriate habitat for proliferation and create host and parasite availability due to nomadic habits and by moving cattle from different areas and concentrating them at places like livestock markets. Through Figure 1, it is possible to view tourist tracks. According to park officials, insecticide (Glocinex®) impregnated tsetse traps and screens are put every 100 meters along those tracks starting October after the early burning exercise and are re-applied every 3 months. This acts as a protective factor (p = 0.0006) making infections more predominant in areas further from the tourist tracks. Kidui has the lowest prevalence (27.08%) and is close to the park and to tourist tracks, Mikumi is also close but its prevalence is 50% probably due to the higher influx of cattle from other areas and due to thicker vegetation found around it.
Access to markets, animal trade and uncontrolled distribution of drugs for veterinary use was also investigated in this study. Results have shown that these human activities strongly influence the occurrence of infection.
When properly applied, chemotherapy along with vector control becomes a very strong method of controlling trypanosomosis [49,50]. In general Diminazene aceturate (Berenil), Homidium chloride (Novidium) and Isometamidium chloride (Samorin) were widely used at the study site. Interestingly, the villages with easier access to these drugs were the ones with higher prevalence of disease, which may suggest parasite resistance due uncontrolled use and misuse of those chemotherapeutic agents. This situation can be troubling if major pathogenic trypanosomes species become resistant to the currently available drugs. Resistant strains of T. congolense and T. vivax have been isolated in Nigeria, Mozambique, Somalia, Zimbabwe and other African countries [51-54]. The measures to combat this nuisance are limited since Africa has a narrow market and insignificant efforts have been made to develop new drugs [55]. Based on this fact it is important to optimize the use of the available drugs to avoid resistance.
The use of insecticides and pesticides on cattle is one of the many tsetse control measures [38]. Although pyrethroids were widely used, their effect on infection occurrence was not clearly evident. In the studied villages insecticide application is done by spraying the animals. This method has negative implications in areas where access to water is limited and the equipment and technique used for their application is not proper leading to inadequate protection of the animals and consequently the people.
A study estimated and mapped human population at risk of sleeping sickness based on reported cases originated from twenty African countries using a Geographic Information System (GIS) [37]. The areas at risk of T. b. rhodesiense infection in Tanzania cover approximately 65,700 km2 and include livestock/wildlife interface areas [37].
In a study done in Uganda spatial analysis of risk factors for HAT was performed and determined that a household with history of infection had higher probability of re-infection and that having a nearby water body was also a significant factor for occurrence of sleeping sickness [35]. Similarly this study shows that human behaviour has a high impact on disease prevalence and that the presence of water bodies also alters disease dynamics. In this case charco dams were shown to be suitable in tsetse infested areas. Being man made water bodies these dams are located close to homesteads and far from vegetation suitable for tsetse fly proliferation. Conditions can be quite harsh especially during the dry season when the charco dams also tend to dry up.
A sleeping sickness outbreak that affected nine people who visited some of the Tanzanian National Parks occurred in 2001 [56]. One of the objectives of this study was to ascertain whether human infective trypanosomes occur in the Mikumi interface areas. However this study did not reveal the presence of T. b. rhodesiense.
Recent data shows a general decline of Human cases of trypanosomosis in African countries [57]. Although reasons for this are not clear the presence of reservoir animals in interface areas, the non-sedentary nature of livestock farmers as well as the arthropod-zoonotic nature of tsetse strongly suggests that HAT is presumably in latency form and hence the reason for its classification as a neglected zoonotic disease.
Prevalence of trypanosome infections was established to be 51.47% in the Mikumi interface areas. Out of all the infected cattle 13.93% presented double infection. Seven trypanosome species were identified in the study area by nested ITS-PCR with Trypanosoma simiae (48.15%) being the most abundant and T. congolense kilifi (0.74%) the least abundant. No human infective T. b. rhodesiense were identified on further SRA-LAMP analysis.
Prevalence of infection significantly varied between villages. The variation of species of trypanosomes among villages relates to the availability of the parasite among wildlife and domestic carriers as well as the existence of cyclical and mechanical vectors in the area.
Dense vegetation and the specific human activities were identified as important risk factors for infection. Areas containing charco dams and those close to insecticide impregnated tsetse traps and screens were significantly associated with a reduction of number of infected cattle. The high prevalence of infection in cattle strongly indicates latency and chronicity rather than clinical disease and this situation poses a constant threat to animal production.
Ethical clearance was granted by the Faculty Research, Publication & Ethics Committee, with the reference number SUA/VET/016/16.
A research permit was granted by the Vice Chancellor of Sokoine University of Agriculture on behalf of Tanzania Commission for Science and Technology (COSTECH).
The study was conducted under the direct supervision of the Kilosa District Veterinary Officer (DVO).
This research came as result of cooperation between Eduardo Mondlane University (Mozambique) and Sokoine University of Agriculture (Tanzania) through a scholarship provided by the INTRA-ACP mobility to fulfill a Masters in Preventive Veterinary Medicine.
The research was sponsored by the OHCEA Grant for Postgraduate Students 2016/2017.
Heartfelt gratefulness is directed to the supervisors of this work, Prof. Kimera and Prof. Gwakisa. Sincere thanks to the laboratory staff in the Genome Science Centre at SUA, Dr. Yuda Mgeni the District Veterinary Officer of Kilosa, to Mr. Peter Koka from the Protection Department of Mikumi National Park and to all who contributed in any way to the accomplishment of this research.