Animal African Trypanosomosis is a Neglected Tropical Disease with significant impacts to pastoral community livelihoods. Our study sought to determine the influence of seasonal cattle movements on the prevalence of trypanosome infections in cattle in the pastoral areas of the Maasai Steppe. Identification of spatial and temporal dynamics of trypanosome infections in cattle is essential for designing effective control strategies. To identify potential hotspots of trypanosome transmission, we worked with 5 pastoralists in each of 3 villages in the Maasai Steppe of northern Tanzania, and consecutively sampled 10 of each of their cattle in 3 periods, covering both wet and dry season grazing ranges (July 2017 to January 2018). Each time blood was collected from the cattle, a prophylactic dose of diminazene aceturate was administered to clear any parasites acquired in the previous 3 months. We then used participatory mapping techniques to identify the areas where the pastoralists had grazed their herds since the last sampling period, and interviewed them about any disease control methods they practice. Trypanosome infections in the sampled cattle blood were detected using nested polymerase chain reaction with ITS-1 primers. The overall prevalence of trypanosome infections across all sample periods and villages was 12%, though this varied distinctly by season and grazing area. Prevalence in July 2017 (19.33%) was significantly higher than prevalence in October 2017 (2%) (p < 0.05). A total of 45 grazing areas were identified and cattle acquired trypanosome infections in almost half of these (n = 21). Targeted awareness on seasonality and hotspot areas of trypanosome infections will help Maasai pastoralists to plan movement of their cattle strategically to avoid disease risk. These results also suggest enhanced control strategies for Trypanosomosis during the months of the year when cattle are moved further from homesteads to graze in hot spot areas.
Cattle movement, Trypanosome prevalence, Maasai steppe, Tanzania, Nagana
AAT: Animal African Trypanosomosis; HAT: Human African Trypanosomiasis; DNA: Deoxyribonucleic Acid; QGIS: Quantum Geographical Information System; PCR: Polymerase Chain Reaction; MCF: Malignant Catarrhal Fever; NM-AIST: Nelson Mandela African Institution of Science and Technology; SUA: Sokoine University of Agriculture; PEHPL: Enhancing Health and Productivity of Livestock; BMGF: Bill & Melinda Gates Foundation
Animal African Trypanosomosis (AAT) is amongst the tropical [1,2] neglected [1], endemic [3] and economically important [4] diseases affecting livestock in many African countries, including Tanzania. The causative agents in cattle include Trypanosoma congolense, T. brucei and T. vivax [5,6]. Other trypanosomes can also be present and impact other domestic animals such as T. simiae which causes nagana in pigs [7] and Trypanosoma evansi which causes surra disease in camels, horses, cattle and buffaloes [8]. In humans, T. brucei rhodesiense (common in East Africa) and T. brucei gambiense (common in West Africa) cause Human African Trypanosomiasis (HAT) and are the etiological agents for sleeping sickness [9,10]. In Tanzania, the disease reduces the socio-economic status of pastoral and agro-pastoral smallholder farming systems. For instance, it has been estimated that 4.4 million livestock and 4 million people in Tanzania are at risk from Trypanosomosis [11]. The total estimated loss per annum in Tanzania from AAT due to mortality and reduced milk yield is estimated at US $ 7.98 million [11]. These effects have triggered many countries, including Tanzania, to develop and enact control strategies such as vector control and treatment of sick animals.
In spite of these control strategies, AAT remains a serious threat to the resource-poor pastoralists in much of Tanzania, including the Maasai livestock herders who occupy the Maasai Steppe in northern Tanzania. As well as village lands used by pastoralists and agro-pastoralists, the Maasai Steppe contains protected areas such as national parks, game reserves and wildlife management areas which host large numbers of wildlife that act as reservoirs of trypanosomes, including eastern blue wildebeest/white-bearded wildebeest (Connochaetes taurinus albojubatus), cape buffalo (Syncerus caffer), common eland (Taurotragus oryx), zebra (Equus quagga) [5,12,13]. The co-existence of livestock and wildlife in the ecosystem is thought to be one of the main factors increasing the vulnerability of cattle to trypanosome infections [14].
Some of the factors that increase risk of AAT in the Maasai Steppe are frequent droughts and shortage of pasture [15] that drive the livestock farmers to move long distances with their cattle in search of water and pasture, and can force them to use areas occupied by wildlife [16,17]. Livestock movements in the Maasai steppe are traditionally controlled by elders of the family. Elders are considered highly knowledgeable in identifying locations that may have suitable pasture with minimal livestock-wildlife interaction risks [18]. Since the available land for cattle grazing has been reduced by conversion to crop farming [17,19] the seasonal livestock movements often lead to encroachment of cattle into protected areas that increases exposure of cattle to tsetse fly bites and trypanosome infections. Tsetse fly abundance in the Maasai Steppe is influenced by vegetation type, climate, distance from protected areas and the availability of wildlife species [14,17]. In Uganda, studies showed that the spread of T. b. rhodesiense into uninfected areas was facilitated by cattle movement during marketing efforts [20,21].
The tsetse fly (Glossina spp.) is the primary vector for trypanosomes. The abundance of the tsetse vector, and the community of trypanosomes they carry and transmit is influenced by habitat factors and host presence [14], as well as time of year [22]. As such, we might expect herders to trade off the availability of good pasture with the risk of exposure to trypanosome infection. In this study we investigated how herders moved their cattle in relation to perceived Trypanosomosis risk by estimating infection rates and prevalence of trypanosome infections in cattle in different seasonal grazing areas. An understanding of disease risk and associated herder behavior can provide insights into the development of grazing management plans aimed at reducing the burden of the disease.
The study was conducted in three villages in the Maasai Steppe: Kimotorok and Sukuro in Simanjiro district, and Oltukai in Monduli district. The three villages were selected because of their differing distances to nearby protected areas, which are considered the main reservoirs of AAT [23] (Figure 1). These villages are found in northern Tanzania, where husbandry practices are predominantly pastoral and agro-pastoral. The pastoralists keep large herds of cattle under extensive management which forces them to move from one location to another in pursuit of pasture and water. The seasonal movements often push the livestock to potential infection hot spot areas which are either agricultural or protected areas where cattle encounter infected tsetse flies. The Maasai Steppe ecosystem covers some 35,000 km2 (9,700 sq mi) [24], spanning from northern Tanzania into southern Kenya (from 3°40' and 4°35' S, to 35°50' and 36°20' E) [14,25]. The area is home to important wildlife populations that move seasonally from protected areas into surrounding community lands, increasing wildlife-livestock interactions. Dominant vegetation types include Acacia-Commiphora woodland, open grassland, seasonal swamps and often dense bushland with dominant tree species including Erythrina burtii, Combretum spp., Albizia spp., and Cordia spp [14]. The area is semi-arid receiving approximately 542 mm of rain per year, with a prolonged dry season from June to October, and a short rainy season from November to January and a long rainy season from March to May [17].
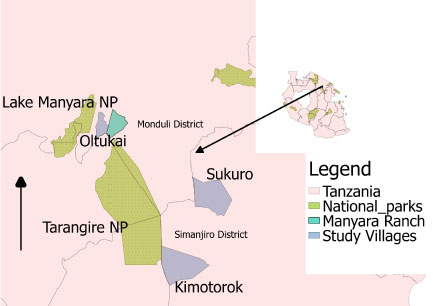 Figure 1: Map showing the three study villages: Sukuro and Kimotorok from Simanjiro District and Oltukai from Monduli district in relation to nearby protected areas.
View Figure 1
Figure 1: Map showing the three study villages: Sukuro and Kimotorok from Simanjiro District and Oltukai from Monduli district in relation to nearby protected areas.
View Figure 1
Sample size was estimated based on prevalence levels in cattle of 17.2% from previous studies in this area [22]. We estimated sample size from the equation: n = (Z2pq)/e2, where n = is required sample size Z = critical value for a given 95% confidence level, e = desired absolute precision = 5%, p = existing prevalence of trypanosome infections in the Maasai Steppe. As such, we needed to acquire at least 219 cattle blood samples to test difference in prevalence between grazing areas.
In each village, 5 bomas were selected randomly from the list provided by the local government leaders. A boma is a Maasai house hold (compound house) complising more than one houses in one place. The houses look like small huts built with mud and cow dung. Heads of households were approached and informed consent sought to sample their cattle. Ten cattle were randomly selected while considering age and sex, however more cows were selected on account of the low availability of bulls in each household. Blood samples were collected in July 2017, October 2017 and January 2018 from 10 tagged cattle in each household. During the first sampling period (July 2017), blood samples were collected and then each animal given a prophylactic dose of diminazene aceturate. Blood re-sampling was done after an interval of three months (i.e. October 2017 and January 2018) and after each sampling the cattle received diminazene aceturate to clear previous infections. Diminazene aceturate becomes ineffective 20-21 days after treatment [5,26] after which it was assumed that the animals could acquire new infections. In case any tagged animal became sick between sampling periods, proper communication was established with the farmers to ensure blood was collected before treating the animal. If an animal was sold or died, replacement cattle were identified to retain sample size of 150 cattle throughout the study. To be included in the study, animals must not have been treated with trypanocidals within last two months. Three milliliters of blood was collected from the jugular vein by venipuncture into an EDTA-containing vacutainer tube from 150 cattle during each sampling session (50 blood samples from each village). The blood was kept on ice and transported to the laboratory where it was kept at -20 ℃ until DNA extraction.
Information pertaining to livestock movement during each trimester was gathered from each of the households in which cattle were sampled. A semi-structured questionnaire was administered to each cattle owner to extract information on patterns and reasons for cattle movement. The pastoralists were requested to indicate on a scale of 1 (most important) to 10 (least important) their reasons for moving their livestock from one location to another. Detailed information on where the cattle had grazed in the previous two months was then acquired using participatory mapping methods. This involved using high resolution satellite imagery from Google Earth Pro (version 7.3.1.4507). Pastoralists were familiarized with known landscape features around their households and villages using maps printed as high resolution images. Google Earth Pro was then used on an Apple Ipad, Version 9.3.5 (13G36) to allow the pastoralists to zoom in and out and better identify known areas, and then use this technique to map where cattle were grazed during the past two months prior to the day of blood sampling. Polygons of the grazing areas were digitized in Google Earth and the shape files exported into Quantum GIS (QGIS Development Team, 2014) for further analysis and creation of grazing maps. After delineating the grazing areas with participatory mapping, each identified area was subsequently visited with the herder for physical validation.
Zymo research kits were used for DNA extraction by following kit instructions provided by the manufacturer (Zymo research, California, USA). Extracted DNA was kept at 4 ℃ until Polymerase chain reaction (PCR). Trypanosome species from extracted DNA were identified using nested PCR technique with specified conditions and reagents shown in Table 1. Two rounds were performed, with 35 cycles each round. The primer sequences used were ITS1: 5'-GAT TAC GTC CCT GCC ATT TG-3'; ITS2: 5'-TTG TTC GCT ATC GGT CTT CC-3'; ITS3: 5'-GGA AGC AAA AGT CGT AAC AAG G-3' and ITS4: 5'-TGT TTT CTT TTC CTC CGC TG-3'. The PCR products were separated on 1.5% GR green stained agarose gels and positive results identified based on PCR products sizes matching to 611 bp for T. vivax, 988 bp for T. theileri, 850 bp for T. simiae, 954 bp for T. simiae tvaso, 1207-1224 bp for T. brucei, 1422 bp for T. congolense, 1413 bp for T. congolense savannah (Figure 2), 1422 bp for T. congolense kilifi and 1513 bp T. congolense forest [27,28]. Gel images were visualized using E-Box imaging system (E-Box CX5 Vilber Lourmart). The images were saved for further analysis to determine the DNA fragment sizes.
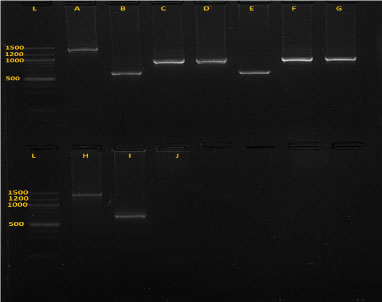 Figure 2: Gel image (picture of the 1.5% agarose gel electrophoresis) showing five species of trypanosomes after amplification.
Figure 2: Gel image (picture of the 1.5% agarose gel electrophoresis) showing five species of trypanosomes after amplification.
A is T .c. savannah (1403 bp), B, E and I are T. vivax (620 bp), C and G are T. thelleri (981 bp), D and F are T. simiae tsavo (954 bp) and H is T.c. kilifi (1422.24 bp), J is negative control and L is ladder.
View Figure 2
Table 1: Target genes for PCR, reagents' volume used for each round and cycling conditions. View Table 1
To test for significance of differences of prevalence among variables (Grazing plots, seasons, villages), we used analysis of variance (ANOVA) for parametric data and Kruskal Wallis for non-parametric data. We used generalized linear mixed model (GLMM) analysis to determine the effect of month of sampling on trypanosome prevalence. The analyses were performed using R statistical software (R Development Core Team 2011).
The overall prevalence of trypanosome infections in cattle across all three sampling periods and all study villages was 12% (n = 450). Prevalence was greatest in July 2017 (19.33%, n = 150), which was significantly higher than in October 2017 (2% prevalence, n = 150) and January 2018 (14%, n = 150) (Kruskal-Wallis Test Statistic = 19.18, P < 0.001, df = 2).
Overall, 8 trypanosome species were detected using the specific nested PCR. T. simiae exhibited the highest prevalence (5.33%) followed by T. vivax (2.89%), whereas the lowest prevalence was found in T.c. savannah (0.22%), T.c. forest (0.22%) and T. brucei (0.22%), with each found in just a single animal (Figure 3). Five co-infections were detected in 5 cattle, 3 being from Kimotorok village and 2 from Sukuro. The 3 co-infections in Kimotorok village were T. vivax and T. theileri; T. vivax and T. simiae and T. vivax and T.c savannah. The 2 co-infections detected in 2 cattle in Sukuro village were T. simiae and T. congolense and T. simiae and T. vivax.
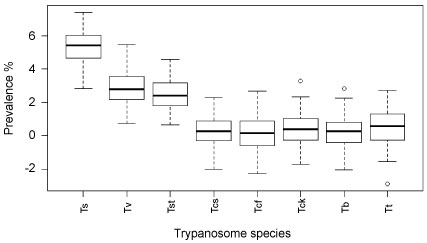 Figure 3: Overall prevalence of trypanosome species (n = 450).
Figure 3: Overall prevalence of trypanosome species (n = 450).
Ts = Trypanosoma simiae, Tst = Trypanosoma simiae tsavo, Tv = Tryapanosoma vivax, Tt = Trypanosoma theileri, Tcs = Trypanosoma congolense savannah, Tb = Trypanosoma brucei, Tcf = Trypanosoma congolense forest, Tcf = Trypanosoma congolense kilifi.
View Figure 3
Seasonal variability was documented for abundance of the trypanosome species, either individually or as co-infections. Thus, 5 trypanosome species, T. simiae, T. simiae tsavo, T. vivax, T. theilleri and T. brucei were detected in July 2017, while only 2 species (T. simiae and T. vivax,) were detected in October 2017, and 6 trypanosome species (T. simiae, T. simiae tsavo, T. vivax, T.c. savannah, T.c. forest, T.c. kilifi) were recorded in January 2018 (Table 2 and Figure 3).
Table 2: Number of cattle carrying trypanosome infections, shown by species. View Table 2
The cattle owners identified a total of 45 grazing plots during the course of this study (Figure 4). Based on information about where the herders had grazed their cattle in the previous 2 months, trypanosome parasites were detected in 21 out of the 45 plots (46.67%). All the cattle belonged to 15 herds and trypanosome parasites were detected in 13 of these at least once during the study period (Table 3, Figure 5a, Figure 5b and Figure 5c).
Table 3: Prevalence of trypanosome infections by village, herd and grazing plot, where n = 10 cows per herd. View Table 3
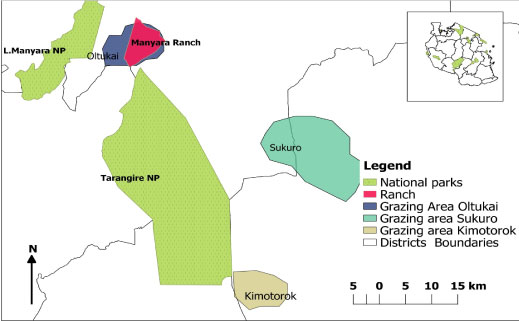 Figure 4: Map showing the study sites and grazing areas in the 3 villages. The overal all grazing area in each village was obtained by combining all the different grazing areas polygons.
View Figure 4
Figure 4: Map showing the study sites and grazing areas in the 3 villages. The overal all grazing area in each village was obtained by combining all the different grazing areas polygons.
View Figure 4
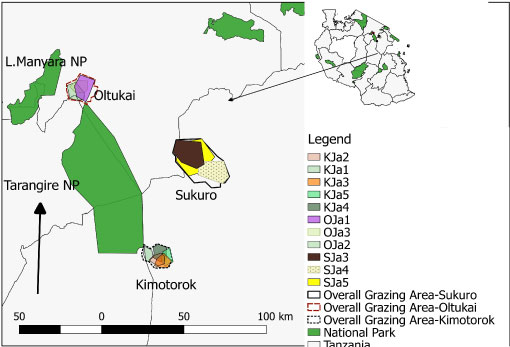 Figure 5A: Map of the grazing areas (plots) in the month of January 2018 for the three villages (Oltukai, Sukuro, Kimotorok) showing percent of infected cattle (n = 10 in each herd during each sampling period).
Figure 5A: Map of the grazing areas (plots) in the month of January 2018 for the three villages (Oltukai, Sukuro, Kimotorok) showing percent of infected cattle (n = 10 in each herd during each sampling period).
(Key: OJa = Oltukai January, SJa = Sukuro January, KJa = Kimotorok January).
View Figure 5A
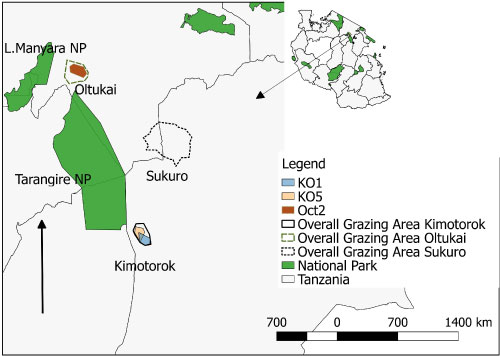 Figure 5B: Map of the grazing areas (plots) in the month of October 2017 for the two study villages (Oltukai, Kimotorok) showing percent of infected cattle (n = 10 in each herd during each sampling period). However, no positive infection was observed in October in Sukuro Village.
Figure 5B: Map of the grazing areas (plots) in the month of October 2017 for the two study villages (Oltukai, Kimotorok) showing percent of infected cattle (n = 10 in each herd during each sampling period). However, no positive infection was observed in October in Sukuro Village.
(Key: Oct = Oltukai October and KO = Kimotorok October).
View Figure 5B
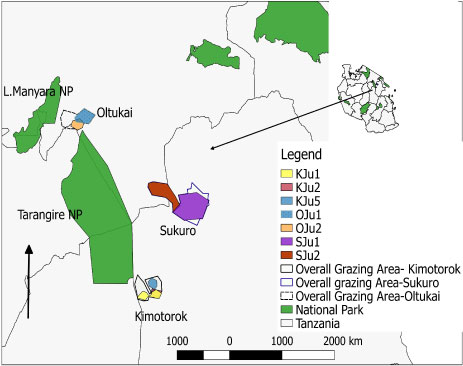 Figure 5C: Map of the grazing areas (plots) in the month of July 2017 for the three study villages (Oltukai, Sukuro, Kimotorok) showing percent of infected cattle (n = 10 in each herd during each sampling period).
Figure 5C: Map of the grazing areas (plots) in the month of July 2017 for the three study villages (Oltukai, Sukuro, Kimotorok) showing percent of infected cattle (n = 10 in each herd during each sampling period).
(Key: OJu = Oltukai July, SJu = Sukuro July and KJu = Kimotork July).
View Figure 5C
Prevalence of the infections varied from one grazing plot to another as well as one herd to another. In July 2017, trypanosomes were detected in cattle from 7 plots (3 in Kimotorok village and 2 each in Sukuro and Oltukai villages) (Table 3, Figure 5a and Figure 5c). In October 2017, trypanosome infections in cattle were detected only from 3 grazing areas (2 overlapping plots in Kimotorok village and one plot in Oltukai village). No infections were detected in Sukuro village in October 2017. In January 2018, trypanosomes were detected in cattle grazing on 5 plots in Kimotorok village and 3 plots each in Sukuro and Oltukai villages.
Overall, parasites were consistently detected in cattle from 2 herds (herds 1 and 5) in Kimotorok village and one herd (herd 7) in Oltukai village. The variations in prevalence of trypanosome infections in cattle between different grazing plots as well as between the 3 sampling periods were statistically significant (F-value = 4.152, P = 0.02, df = 2). Furthermore, GLMM analysis identified the highest infection during July 2017 and the variation in prevalence of trypanosomes in relation to grazing plots was lower in October 2017 and January 2018 (Table 4).
Table 4: The association of trypanosome prevalence with month of sampling, where July is the intercept. View Table 4
Herders were asked to rank the reasons for moving their cattle herds from one area to another. Their responses are shown in Table 5. Search for pasture and water during the dry season was the most frequently mentioned reason for cattle movement, whereas avoidance of invasive plant species and ownership of large numbers of cattle were least mentioned by the livestock owners. Some variables were equally scored by respondents across the 3 villages.
Table 5: The mean rank scores of drivers for cattle movement. The ranking is based on a sample size of 45 respondents, 15 in each village. View Table 5
The response and opinions of individual farmers differed when responding to questions about different drivers for cattle movement. For instance, higher response on search for pasture was observed in Sukuro and Kimotorok while search for water was reported to be highest in Kimotorok (Table 5).
The ranking on the reasons for animal movement varied among villages. In All the three villages search for pasture scored highest followed by search for water. When ranking in diseases avoidance was compared among the three villages, the highest score was observed in Oltukai (Table 5). However, the overall variation of the mean scores between the three villages was insignificant (Kruskal Wallis = 0.62, p = 0.2, df = 2).
We further examined the difference in prevalence of trypanosome infections in cattle owned by pastoralists who moved their herds to avoid disease, compared to those who did not. In this case, the disease the pastoralists were avoiding was Malignant Catarrhal Fever (MCF), which is transmitted in the after birth of wildebeest. Pastoralists practicing disease avoidance move their cattle away from the plains where the wildebeest calve, which could force them to use areas with greater trypanosome infection risk. In fact, our results showed significantly higher prevalence of trypanosome infections in cattle when the herders practice disease avoidance of MCF (Mann-Whitney U test statistic = 17559.500, P < 0.0001, df = 1) (Figure 6).
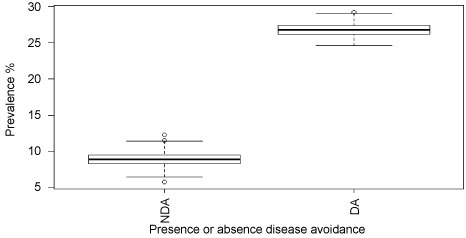 Figure 6: Prevalence of trypanosome infections when the pastoralists are practicing or not practicing avoidance of disease (MCF).
Figure 6: Prevalence of trypanosome infections when the pastoralists are practicing or not practicing avoidance of disease (MCF).
Key: NDA = Not practicing diseases avoidance, DA = Practicing disease avoidance.
View Figure 6
The aim of this study was to examine the influence of cattle movements on trypanosome infection rates, and in particular to identify seasonal hotspots of infection risk in grazing areas used by 3 different villages in the Maasai Steppe of northern Tanzania. Our results revealed a mean infection rate of 12%, over all 3 sampling periods in July and October of 2017 and January 2018. The mean prevalence established in this study is comparable to other reports conducted in northern Tanzania, in which [22] found a 17.2% prevalence in the same areas of the Maasai Steppe. However, others have reported slightly higher or lower prevalence than this. For instance, [7] reported a lower prevalence of 4.2-5.8% while [29] reported a higher prevalence of 51.47%. Differences in climate and ecological factors together with sampling and detection methodologies may explain the differences of the results from the various studies. For instance, a study by [29] which showed higher prevalence of trypanosome infection in cattle included Esilale villages which is known to vary in term of wildlife abundance compared to Kimotorok, Sukuro and Oltukai.
Infections in our study were from 8 different trypanosome species, of which T. simiae was the most prevalent. This agrees with [4] who reported this species to be most prevalent in another area with a wildlife-livestock interface in eastern Tanzania. Besides high prevalence of T. simiae, presence of T. vivax (2.89%) and T. congolense (0.22%). The infection rate for T. vivax was 4.65% in January for Kimotorok. While infection rate being 6.12%, 2% and 7.5% in July, October and January respectively for Oltukai, 3.9% and 4.89% in July and January respectively in Sukuro. The infection rate for T. congolense was 1.96% in July for Kimototok. This is of more interest in this study considering that these species are pathogenic to cattle [29,30], regardless of their lower prevalence. The finding of pathogenic species of trypanosomes in the study area is also of relevance to pastoralists, as it confirms risk and vulnerability of their cattle to the disease AAT.
We showed that prevalence of trypanosome infections varied between months of the year, being highest in July followed by January. These results suggest presence of at least two peaks of infection prevalence in a year, July and January. These months of the year correspond to the dry period in the Maasai Steppe. July as well as January are dry months of the year; accordingly, shortage of pastures on grazing lands and drought are expected to lead to nutritional stress and decreased immunity of the cattle, making them vulnerable to Trypanosomosis as was noted by other researchers [6,31-33]. Concurrently, lower trypanosome prevalence was indicated in the month of October. A plausible explanation to this result relates to our previous study done in the same villages, whereby we showed that prevalence of trypanosomes in tsetse flies increased from January to November with some monthly fluctuations [17]. The presence of trypanosomes in cattle throughout the year in our study area could be associated with several factors including: the year-round presence of wild animals as "trypanosome reservoirs", infestation of tsetse flies (vectors) [14,34] and the frequent cattle-wildlife interaction in common grazing area, thus allowing the chance of year-round trypanosome circulation in the area, [14]. The low trypanosome prevalence recorded in October was unexpected but this was possibly due to the fact that October coincides with a period of short rains when livestock graze on plots close to homesteads, thus reducing rates of contact between cattle and vectors. Also, low cattle-wildlife interactions shairng similar grazing areas during this season due to most of wildlife being back into TNP.
The prevalence of trypanosome infections varied between months of the year (season) as well as between grazing areas in the three study villages. The results are important to identify specific areas where cattle are likely to pick new infections as they move from one grazing location to another in search of pasture and water. We found more infections in grazing plots located in Kimotorok village than in Sukuro and Oltukai villages. Herders in Kimotorok village normally graze their cattle in pastures adjacent to Tarangire National Park (TNP), especially during the dry months of the year. Not surprisingly therefore, the highest number of infections was observed in the month of July, a time when herders encroach areas bordering TNP with their livestock for pasture and water. This is in agreement with a study done in this area which revealed that abundance of tsetse flies and host species increased adjacent to TNP [14,17]. Similar findings were observed by [4] that proximity to Mikumi National Park had implication on prevalence of trypanosome infection in cattle.
Because pastoralists depend on their livestock to meet their livelihood needs, and AAT has significant negative impacts on livestock production and therefore human health, it is essential to understand the dynamics of the disease as it relates to cattle movemements. Such an understanding can help identify hotspots of infection and guide decisions on controlled livestock movement and rational utilization of grazing areas during prolonged dry seasons. Based on our survey, it became evident that the major driver forcing Maasai pastoralists to move their livestock from one area to another is pursuit for pasture and water, regardless of what the risk of AAT might be. Other drivers for cattle movement are avoidance of cultivated areas to minimise conflicts with crop farmers and avoidance of Malignant Catarrhal Fever (MCF) disease-prone areas, for example calving grounds used by wildebeests known to carry risk of Malignant Catarrhal Fever (MCF) infection [35]. However, inspite of efforts by pastoral community- using diseases (MCF) avoidance as strategies to rescuer their livestock, yet cattle become vulnerable to trypanosome infections as they continuously encounter tsetse fly bites in new grazing areas.
Our results are based on a small sample size and limited to 3 villages. However, the results demonstrate the factors that maintain the problem of a neglected tropical disease, Animal African Trypanosomosis, among communities of resource-poor pastoralists in an endemic setting. Understanding the temporal and spatial dynamics of potential hotspot areas in the ecosystem can help pastoralists make decisions about cattle movement patterns to reduce risk in areas where infection is mostly likely to occur.
Seasonal cattle movements have a significant impact on the prevalence of trypanosome infections in cattle in the Maasai Steppe. Time of the year and grazing in areas close to the wildlife interface have been shown to influence prevalence of trypanosome infections in cattle. Based on farmers' opinions, the major driver for movement of their livestock is search for pasture and water, and this is exacerbated during the dry season. These results imply that coordinated planning of grazing patterns may be necessary to reduce risk of infection in such areas as the Maasai Steppe, where AAT is endemic. Also, educational programmes to pastoralists on control strategies for Trypanosomosis including controlled livestock movement and increasing grazing reserves in areas farther from the wildlife interface can reduce the severity of this problem. Furthermore, treatment effort to diseases cattle and tsetse control effort such as tsetse flies trapping, spraying and Insecticide-impregnation of lower limbs to cattle should be emphasized when cattle are grazing in wildlife-cattle interaction zones which are well known to be risk areas.
This research was supported by the Programme for Enhancing Health and Productivity of Livestock (PEHPL) funded by the Bill & Melinda Gates Foundation (B&MGF).
The authors declared that they have no competing interests.
Human and animal ethical approvals were obtained from the re-search ethics committee at the Nelson Mandela African Institution of Science and Technology. The research Permit was provided by the Executive directors from Simanjiro and Monduli Districts council. Informed consent forms (ICF) were filled by the pastoralists to whom the data were collected.
The Programme for Enhancing Health and Productivity of Livestock (PEHPL) funded by the Bill & Melinda Gates Foundation (BMGF) is highly appreciated for providing financial support. We would like to give thanks to the following field work team members who were very supportive during sample and data collection: Oleshongoni Boniphace, Edward Kumbi, Mokia Mirimbi and Ukweli Kidonya. We are cheerfully appreciating the laboratory technical support provided by Asteria Butuyuyu and Fatuma Kindoro from Genome Science center at Sokoine University of Agriculture (SUA). Furthermore, we are thankful to all staff at Nelson Mandela African Institution of Science and Technology (NM-AIST) for the positive co-operation rendered during the PhD training of the first author.