Not only O157 Shigatoxin-producing Escherichia coli (STEC) but some of non-O157 STECs are attracting attentions as clinically important STEC and a possible food-borne spread of these pathogens is becoming a worldwide concern. In this study, we developed a sensitive, specific, and simple method to detect STECs of clinical significance in retail beef so that it can be used even in resource-limited countries. We designed and fine-tuned a protocol characterized by PickPen device-assisted immunomagnetic separation (IMS) for specific and simultaneous screening of O157 and eleven other O serotypes and quick and easy stx gene detection in STEC by the loop-mediated isothermal amplification (LAMP) assay. In vitro experiments showed 91CFU/ml sensitivity for IMS and 100% specificity for the IMS-LAMP-based method. We detected by the IMS-LAMP-based method the stx gene in 41 of 74 (55.4%) and none of 28 (0%) of beef purchased in, respectively, Thailand and Japan. Analyses of the STEC isolates from the Thai beef demonstrated high sensitivity and high (80-100 %) specificity of the detection method and regional difference in the STEC distribution in beef; and therefore the results support that this IMS-LAMP-based method is suitable for detection of clinically important STEC in beef.
Shigatoxin-producing Escherichia coli, Retail beef, Immunomagnetic separation, PickPen, loop-mediated isothermal amplification
STEC: Shigatoxin-Producing Escherichia coli, EHEC: Enterohaemorrhagic Escherichia coli, stx: Shigatoxin gene, stx1: Shigatoxin gene type 1, stx2: Shigatoxin gene type 2, IMS: Immunomagnetic Separation, IMB: Immunomagnetic Beads, IMBv: Immunomagnetic Beads Volume, bIMBv: Best Immunomagnetic Beads Volume, LAMP: Loop-Mediated Isothermal Amplification, CE: Capture Efficiency
Shiga toxin (Stx)-producing Escherichia coli (STEC; also called verocytotoxin-producing E. coli or VTEC) can cause mild diarrhea in humans. Enterohaemorrhagic E. coli (EHEC) is a highly virulent sub-group of STEC and is one of major categories of pathogenic E. coli that cause important enteric infections in humans [1]. The common virulence factor of STEC and EHEC is the production of one or both subtypes of Stx (Stx1 or Stx2). EHEC is defined, based on the typical symptom of the patient, as STEC causing severe bloody diarrhea in humans. The EHEC may be confirmed experimentally by detection of additional virulence factors intimin and the large EHEC plasmid [2].
Strains of STEC/EHEC are differentiated by the O antigen on their outer membrane into over 180 serotypes [3] Of all EHEC serotypes E. coli O157:H7 is the one which has caused most food borne outbreaks and has led to the highest number of hemolytic uremic syndrome cases, a life-threatening sequela of STEC infection; however, some of non-O157 serotypes are also becoming an important health concern because they have been isolated from the patients with the typical symptoms of EHEC infection with increasing frequency than before [4,5]. EHEC belonging to O157 and six other O serotypes (O26, O91, O103, O111, O121, and O145) are considered important in Japan [6-8]; and additional eight serotypes (O15, O45, O55, O104, O113, O118, O128, and O153) totaling to fifteen O serotypes for all countries in the world [9-15]. We therefore considered these fifteen O serotypes of EHEC are of particular importance for a global-scale detection method.
In the inspection of environmental samples including food, the STEC expressing the known O antigens of EHEC are targeted as a potential public health hazard. Serogroup-specific method and virulence-specific method were combined for detection of STEC belonging to a clinically important O serogroup in some studies. Combinations of simple to sophisticated and high-cost techniques targeting a narrow spectrum of the important O serotypes of STEC/EHEC (a maximum number of eight of target O serotypes). These include multiplex PCR [16], PCR-ELISA and multiplex real-time PCR assays [17] and LAMP [18]; multiplex real-time PCR assays screening for virulence genes and O-antigens encoding genes [15]; immunomagnetic separation (IMS) targeting certain O serotypes of E. coli individually or simultaneously [19,20]; and a combination of multiplex real-time PCR and IMS assays [21-23].
Due to the rapid development of transportation, the food trade among various countries has become very active these days. Therefore, specific, sensitive and easy-to-perform methods are necessary for examination of food for a wider range of the important O-serotypes of STEC/EHEC even in resource-limited and tropical countries. In this study, we combined a unique IMS method, as a specific and easy-to-perform, covering clinically important O serotypes worldwide and a loop-mediated isothermal amplification (LAMP) method [24], as a simple, sensitive and low-cost technique, to achieve the goal described above.
Magnetic beads were coated with antibodies against seven E. coli O antigens using antibodies partially purified from rabbit polyclonal antiserum specific to each of target O antigen (rabbit anti-Escherichia coli O157, O26, O91, O103, O111, O121 or O145 antiserum, Denka Seiken Co. Ltd., Tokyo, Japan). These resulted in bead group 1 suspension that targets the O serotypes of clinical importance in Japan. Similarly, bead group 2 suspension containing the beads coated with antibodies against anti- O15, O55, O104, O128 and O153 was prepared. Antibodies against anti- O45, O113 and O118 were unavailable for us and thus were not included. Equal volumes of bead group 1 suspension and bead group 2 suspension were mixed, resulting in the final bead suspension and designated as IMB in this study. This bead suspension (IMB) targets the O serotypes of clinical significance on a global scale. IMB was stored at 4o C and used whenever needed throughout this study.
Japanese beef was purchased from 4 local stores in Kyoto City, Japan, in 2013. Malaysian and Thai beef were purchased from a local market in Hat Yai City, southern Thailand, in 2013 and 2014. The protocol used to examine the beef samples is outlined in Figure 1. Twenty five grams of each sample was incubated at 37°C in 225ml of tryptic soy broth (Bacto™ Tryptic Soy Broth; Difco, Becton Dickinson Microbiology Systems, Sparks, MD, USA). After 6 h, 1 ml of the culture was added to 250ml of modified EC medium containing 25 mg/l novobicin (Nobobiotin added m-EC medium, Kyokuto Pharmaceutical Industrial Co., Ltd., Tokyo, Japan) and incubated at 42o C for 18 h. The broth culture with or without IMS treatment was examined in two steps. Step 1: the DNA template was prepared by a boiling method: boiling 1ml of the broth culture or bead suspension for 10 min, chilling on ice for 10 min, followed by centrifugation at 15,000 rpm for 5 min and collection of the supernatant. The DNA template was examined for the presence or absence of the stx genes by a conventional PCR method as previously described in [25] and the LAMP method according to the instruction by the manufacturer (Loopamp™ Verotoxin-producing Escherichia coli Detection Kit, Eiken Chemical Co., Ltd., Tokyo, Japan). Step 2: ten µl of the broth culture or bead suspension was streaked onto two selective agar media and incubated at 37°C for 18 h. Up to 5 suspected colonies from each triplicated agar plate were selected: mauve colonies growing on CHROMagarTM STEC (with 50 mg/l STEC supplement) and blue colonies growing on CHROMagarTM ECC (CHROMagar, Paris, France). The selected colonies were grown in 2 ml of Luria-Bertani broth [26] and the DNA template was prepared and examined by conventional PCR for stx1 and stx2 genes. The isolates that gave stx1+ and/or stx2+ result were examined for the eae gene as previously described in [27] and for O serotype by an agglutination test using E. coli O antisera (E. coli antisera “SEIKEN”, Set 1, Denka Seiken Co., Ltd.).
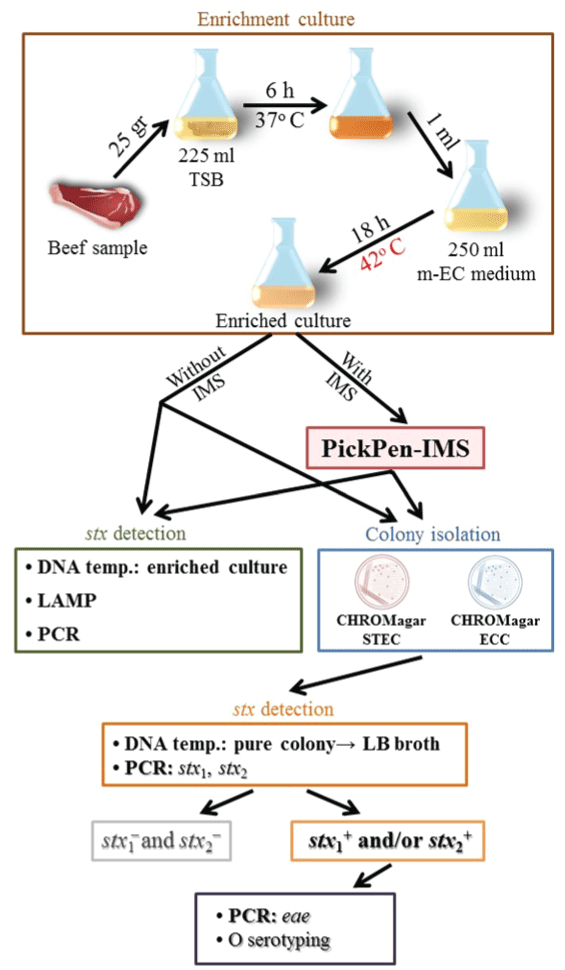 Figure 1: Schematic representation of the protocol for PickPen assisted IMSLAMP-
based detection method for STEC in beef samples.
View Figure 1
Figure 1: Schematic representation of the protocol for PickPen assisted IMSLAMP-
based detection method for STEC in beef samples.
View Figure 1
A 96-deep well (2-ml capacity) titer plate–based IMS procedure using an eight-channel PickPen device (PickPen® 8-M, BIOCONTROL, Washington, USA) was performed as follows. IMB were placed at the room temperature on a shaker (140 rpm) for 30 min before use. Thirty µl of IMB were mixed with 1 ml of the broth culture in the first well and incubated at the room temperature on a shaker (140 rpm) for 30 min. The PickPen device was applied for 5 min and the captured IMB were transferred to the second well and washed in 1-ml washing buffer [phosphate-buffered saline consisting of 0.01M phosphate and 0.16M NaCl with pH 7.0 (PBS) with added 0.1 % Tween 20]. The washing step was repeated in the third well. Finally, the washed IMB were suspended in 1 ml of the washing buffer in the fourth well (designated as Yeild).
The capture efficiency (CE) % was selected as indicator to evaluate four factors (IMB volume hereinafter referred to as IMBv, incubation time of the IMB-cell suspension mixture, PickPen application time, and washing step) in IMS sensitivity test. E. coli PV10-104 was used in this experiment (Table 1). The strain was grown in Luria-Bertani broth at 37o C for 18 h with shaking (160 rpm). Serial 10-fold dilutions of the culture were made in PBS without Tween 20.
Table 1: Reference strains of E. coli used in IMS sensitivity and specificity experiments. View Table 1
IMBv: one milliliter of cell suspension of each of 10-5, 10-6 and 10-7 dilutions was mixed with (20 to 90 µl) of IMBv. The best IMB volume (bIMBv) was determined.
IMB-target incubation time: four incubation times (5, 15, 30, and 45 min) were tested by incubating 10-5 cell suspension of the target cell with bIMBv.
PickPen application time: CE % at selected time points (0.5, 1.5, 3.5, 5, 7 min) of the PickPen application (duration of attracting the IMB by the PickPen) was determined as described below. The CE % relative to the increase in PickPen application time was calculated and the appropriate PickPen application time was determined.
Washing step: 10-5 target cell suspension was incubated with the bIMBv for 30 min. The IMB-target cell complex was washed twice and the CE % was determined and the loss of the target cell was calculated.
The CE was calculated as previously described, CE (%) = (C0 - Ca)/C0 ×100, where C0 is the total number of cells present in the sample (CFU/ml), and Ca is the number of cells not bound to IMB (CFU/ml) [28].
The IMS sensitivity (CFU/ml) was defined as the minimum concentration of target cells detectable by IMB at the bIMBv. This was calculated as follows, IMS sensitivity (CFU/ml) = (CL × CE)/100, where CL is the low concentration of the examined cell suspensions.
To evaluate the specificity of the PickPen-IMS used in this study a mixture of E. coli strains consisting 6 strains each of targeted and non-targeted O serotypes was treated by this method. After the PickPen-IMS treatment the O serotypes and stx genotypes of the E. coli remaining in the solution in each deep well of the 96-deep well titer plate were analyzed relative to those of the starting mixture. The details of the experiment are as follows. The twelve E. coli strains (Table 1) were grown in Luria-Bertani broth at 37°C for 18h with shaking (160 rpm) and the cultures were diluted to 10-5CFU/ml. Equal volumes of each bacterial suspension were mixed in one tube (designated as the starting mix). 20 µl of the starting mix was spread on CHROMagar™ ECC in triplicate to screen the starting composition of O serotypes without IMS (as a control). 1 ml of the starting mix was transferred to the first well and mixed with bIMBv, and then PickPen-IMS was performed as described above. The beads were washed in the second and the third wells and finally suspended in the fourth well. Appropriate volumes of the remaining solution in each well were spread on CHROMagar™ ECC in triplicates and incubated at 37°C for 18 h (20, 50, 50, and 20 µl, from the first to the fourth well, respectively). Colonies were picked up randomly (up to 24 per plate) and the O serotypes were examined for the selected colonies. DNA templates were prepared from the staring mix and from the solutions remaining in the four wells by boiling and examined by LAMP as described above.
All experiments were conducted in triplicate. The means and standard deviations of all collected data were calculated from independent group of replicates. A Student's t-test was used for statistical analysis of the data between two groups. A p-value <0.1 was considered statistically significant.
In this study, we aimed at developing a specific, sensitive and easy-to-perform detection method for clinically important O serotypes of STEC that can be used in all countries in the world. We examined retailed beef samples (not artificially contaminated) by the procedure outlined in Figure 1 to evaluate our IMS-LAMP based detection method. We conducted our investigation in Japan and Thailand where distribution of the target bacterium is relatively scarce and prevalent respectively [25,33-35]. In our strategy, initial screening utilized detection of the stx gene. We examined not only the boiled supernatant of the enriched culture but also isolated colonies for the stx gene in order to check the possibility of false positive and false negative result of the LAMP assay. Twenty-eight Japanese beef samples were examined in 2013. And a total of 74 beef samples (22 and 52 samples, respectively, in 2013 and 2014) purchased in the market in southern Thailand were examined. All 28 Japanese beef samples examined were negative for the stx gene with or without IMS treatment. The Japanese result is largely reflecting the high quality and hygienic conditions of beef production in Japan.
The result of the examination for the presence of the stx gene in the enriched culture of the 74 beef samples purchased in Thailand was classified into patterns from A to F. This classification is based on the difference in the result obtained by the two genetic detection methods (LAMP and PCR) of the enriched culture treated without or with IMS. Forty-eight (64.9 %) of the 74 samples purchased in southern Thailand gave stx+ in at least one result of the four categories when enriched beef cultures treated with or without IMS were examined by LAMP and PCR (Table 2). The total number of stx+ DNA samples obtained by LAMP assay was 26 times (78/3 DNA samples) higher than that obtained by conventional PCR assay.
Table 2: The result of examination of DNA from enriched culture of beef samples for the stx gene purchased in Thailand. View Table 2
The 74 beef samples consisted of 35 Thai beef samples and 39 Malay beef samples. 57.1% of the Thai beef and 71.8% of the Malay beef samples gave stx+ result (data not shown). This result supports the previous report that Malay beef is more frequently contaminated by STEC than Thai beef [33].
In the study in Thailand, we observed two false negative cases of LAMP assay; where LAMP assays gave negative but some isolated colonies gave stx+ result (Tables 2,3, Pattern D, AYK4-1, AYK4-2, AYK4-3 and AYK7-1, LAMP without IMS). The result may suggest the need for a different DNA template preparation method for the LAMP assay other than the boiling method.
Table 3: Characteristics and isolation procedure and origin of the STEC strains isolated in this study. View Table 3
The beef samples classified to Patterns C and D amounted to 39 (52.7 %) beef samples that gave positive result only by LAMP but not by PCR regardless of IMS although stx+ result could be confirmed for 4 (14.3 %) and 4 (36.4 %) beef samples of Patterns C and D, respectively, by obtaining stx+ isolates. This result demonstrated how LAMP is more sensitive than PCR and that PCR failed in detecting the stx gene in the 8 (Patterns C and D combined) beef samples confirmed by isolation of the stx+ strains (false negative PCR result). The Pattern D is composed of 11 (14.9 %) samples that gave stx+ result by LAMP only when IMS treatment was applied, indicating the improvement of the sensitivity by IMS treatment prior to LAMP assay.
However, the Pattern B consists of 7 samples that were positive by LAMP without IMS and turned into negative by LAMP with IMS and we could not isolate any STEC strain which can be explained by a possibility that these 7 samples were contaminated with non-target E. coli O serotypes or non- E. coli bacteria possessing the stx gene [36]. We suspect that the LAMP positive result in Pattern B is most likely to be false positive.
To our surprise, one sample showed Pattern F where STEC O103, one of our target O serotypes, was isolated without IMS but not with IMS; and this particular strain was isolated on CHROMagar ECC only but not on CHROMagar STEC (Tables 2 and 3, AYK6-1). A possible explanation for the first unexpected observation that no STEC was isolated after IMS treatment is that the sample contained a large number of competing populations (E. coli expressing target O serotypes but lacking the stx gene). Because of abundance of competing population, the total number of STEC O103 after IMS treatment probably became below the detection limit of the PCR assay but was still detectable by the LAMP assay. To explain the second unexpected observation that the strain isolated on CHROMagar ECC was not isolated on CHROMagar STEC (contains potassium tellurite), we investigated the possibility that the isolated strain may lack the tellurite resistance gene (terB) [15]. We confirmed this was the case by the PCR assay (Supplement 1).
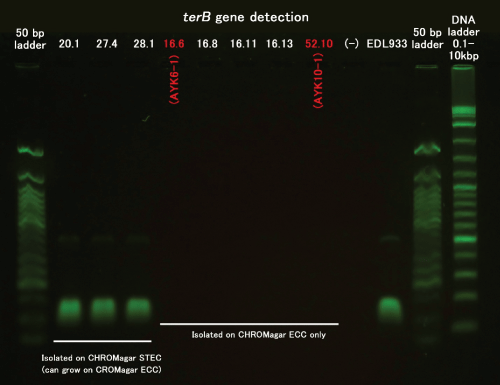 Supplement 1: 16.6: AYK6-1
Supplement 1: 16.6: AYK6-1
52.10: AYK10-1.
View Figure 3
The selective isolation of STEC through CHROMagar ECC but not CHROMagar STEC was also observed in another case (Table 3, AYK10-1) which was also confirmed to be terB- Our result summarized in Table 3 showed that 10 of 12 (83.3%) STEC strains were isolated on CHROMagar STEC but that 2 of 12 (16.7%) STEC strains could be isolated only on CHROMagar ECC. This result is similar to the report by Tzschoppe et al. [15] where 13.6% of EHEC and EHEC-like strains did not grow on CHROMagar STEC. We therefore propose simultaneous use of CHROMagar ECC for the isolation of STEC/EHEC.
In general, we could isolate stx+ E. coli strains more often from CHROMagar STEC (83.3 %, medium S in Table 3) compared to CHROMagar ECC (16.7 %, medium E in Table 3). Twelve strains belonging to six O serotypes were isolated from 10 beef samples. Nine and one of the ten beef samples were imports from Malaysia into Thailand and domestic product of Thailand, respectively. Of the six O serotypes three (O157, O111 and O103) belonged to the O serotypes of IMS target; two strains of O157 were obtained only with IMS and two strains of O111 and five strains of O103 were obtained with or without IMS. The remaining three O serotypes (O71, O140 and O144) were IMS non-target O serotypes; one strain each of these serotypes was obtained exclusively without IMS. The isolated strains included two O157 strains carrying the Stx2 and eae genes; and the other non-O157 strains sharing the Stx1 gene and lacking the eae gene (Table 3).
There are the studies reporting isolation of STEC/EHEC belonging to O157 serotype, possessing the Stx2 gene, but producing no or low-level Stx2 from the beef marketed in Hat Yai, southern Thailand [25,33,35]. Sukhumungoon et al. [35] also reported a non-O157 strain carrying the Stx1 gene from the beef purchased in the same mentioned city in Thailand. Their Stx1+ non-O157 strain produced a large amount of Stx1. In our study carried out in the same city, we isolated 10 strains of non-O157 STEC and all carried the Stx1 gene. Since this type of non-O157 STEC seems to be prevalent in the beef marketed in this area they may be of public health significance.
The result of the analysis of retail beef samples showed that we could isolate the strains carrying the stx gene from 20.8 % of the stx+ samples. We explored by in-vitro experiments the possibilities that IMS-LAMP performance can be further improved by fine tuning the factors involved in the IMS. The following four factors were evaluated in details using CE % as the indicator.
IMBv: incubation time of IMB-target cell mixture (30 min) and PickPen application time (5 min) were fixed. The effect of the combination of various concentrations of the target cells (10.5×101, 10.5×102, and10.5×103 CFU/ml) and the various IMBv (20, 30, 40, 50, 60, 70, 80, and 90 µl) were compared (Figure 2). The average of the CE % was gradually increased and its standard error was reduced with increase in the IMBv. For comprehensive evaluation of IMBv effect, when the result of CE % of the three concentrations of the target cells were combined, the average ± standard error increased from 25.0 ± 47.3 CE % at 20 µl IMBv to 88.4 ± 5.71 CE % at 80 µl IMBv. However, increase in the IMBv from 80 to 90µl had no significant effect on the increase of CE % whereas IMBv increase from 70 to 80µl showed significantly increase CE % for the middle and high concentrations of target cell suspension (p=0.1). Therefore, the 80 µl of IMBv was judged to be the bIMBv. We found, when IMBv was increased to 80 µl or above, the CE value reached a plateau regardless of the cell concentration. Since the ability of IMB to capture the maximum amount of the target cells is limited by the amount of antibodies coating the beads we judged the IMBv of 80 µl (bIMBv) is the most suitable for the studies where the concentration of the target cells is unpredictable. The 80 µl of IMBv is much more than the volume reported by other workers (20 µl) [21,37] and close to the maximum volume that can be handled by the PickPen device in our hands.
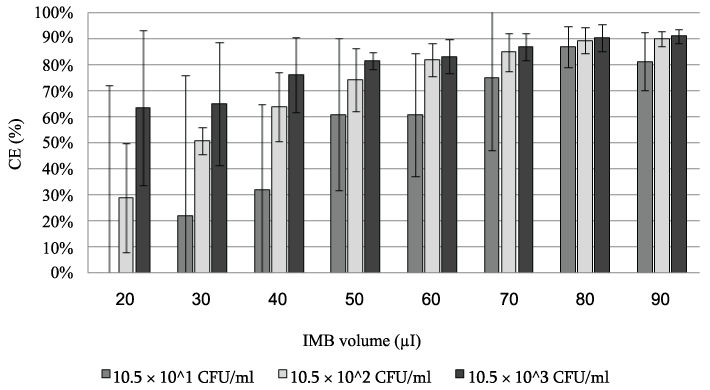 Figure 2: The CE % obtained from three suspensions of the target cell using
various volumes of IMB.
View Figure 2
Figure 2: The CE % obtained from three suspensions of the target cell using
various volumes of IMB.
View Figure 2
Incubation time of IMB-target cell: 2.7×103CFU/ml of the target cell incubated with 80 µl of IMB were employed. The result (5 min, 13.3 ± 3.5 CE %; 15 min, 70.9 ± 6.9 CE %; 30 min, 90.2 ± 0.4 CE %; 45 min, 86.1 ± 5.1 CE %) showed that the CE % was increased significantly when the incubation time was increased from 5 to 15 min and from 15 to 30 min, but not from 30 to 45 min (p = 0.1). Thus, 30 min incubation time was judged as the required time for the best CE % result.
PickPen application time (duration of attracting the IMB by the PickPen): 1.1×103 CFU/ml of the target cell incubated with 80 µl (bIMBv) of IMB were employed. The results (time - CE) were as follows: 0.5 min - 62.1 ± 30.6 CE %; 1.5 min - 57.6 ± 21.7 CE %; 3.5 min - 65.8 ± 11.6 CE %; 5 min - 83.0 ± 4.73 CE %; 7 min - 61.5 ± 14.2 CE %. P values between the neighboring two time points were: p = 0.44, 0.5 - 1.5 min; p=0.24, 0.5 - 3.5 min; p=0.08, 3.5 - 5 min; p=0.08, 5 - 7 min. The p value was gradually reduced as the time increased, but it appeared to have reached a plateau at 5 min. We therefore judged 5 min is the appropriate application time of PickPen.
Washing step on the loss of IMB-target cell complex in the buffer: the first and the second washing had insignificant effect (p = 0.1) (< 1 CE % and < 2 CE %, respectively).
After fine tuning of the IMS factors were fixed to as follows, the IMBv to 80µl, incubation time to 30 min, PickPen application time to 5 min and washing step to be repeated twice. These conditions determined the sensitivity of the IMS as 91 CFU/ml at the low concentration of the target cell (Figure 2).
To evaluate the specificity of the IMS method in vitro, 6 strains of E. coli carrying the Stx1 and/or the Stx2 genes, each expressing different target O antigens, were mixed with 6 strains of E. coli lacking the stx gene and each expressing non-target O antigens. After completion of the IMS treatment, distributions of non-target and target O serotypes in the starting mix (not treated with IMS) and solutions or the cell suspension remaining in the four wells after IMS treatment were examined (Table 4). The result showed that IMS helped to separate and concentrate all target O serotypes (Table 4, Yield). The transition of the change in percentages of target vs. non-target groups showed how and to what extent each step of the PickPen-IMS treatment could contribute to change in the composition of the starting mix. The examination of DNA templates of the 5 suspensions by the commercially available loopamp VT E. coli detection kit reflected how LAMP can perform with the help of IMS in this particular experiment as follows: without IMS, no wash, first wash, second wash, and yield, showed respectively, positive, positive, negative, negative, and positive result for the stx gene. This result suggests that the remaining target cells in the solutions, first wash and second wash, were below the detection limit of the loopamp kit (60 CFU/test).
Table 4: O serotype-based grouping and the number of the colonies randomly selected from the agar plates inoculated with the starting mix (without IMS) and the solution or the suspension remaining in each of the four wells after PickPen-IMS treatment (with IMS). View Table 4
We have evaluated the specificity of the IMS by an in vivo assay (Table 3). In the in vivo study of the beef purchased in Thailand, STEC strains belonging to the target and non-target O serotypes were isolated from 8 and 2 LAMP-positive beef samples, respectively. These non-target O serotypes were O140 and O144 (strains AYK9-1 and AYK10-1, respectively, in Table 3), and they were isolated from two different beef samples only without IMS treatment (Table 2, Pattern C). The LAMP positive result with IMS treatment of these samples may or may not be due to these non-target O serotypes. Based on this result, the specificity of the IMS-LAMP detection method can be judged to be very good ranging from 80 % (due to non-target O serotypes) to 100 % (due to target O serotypes). In addition, the IMS was shown to be 100 % specific by an in vitro assay where only target O serotypes were isolated from the Yield (Table 4).
In our current study we designed an IMS-LAMP method targeting as many as twelve O serotypes and to the best of our knowledge, this is the first time to report this number of O serotypes of STEC targeted at once. We have evaluated the specificity of this method by in vivo and in vitro assays and it showed to be 80 to 100 % specific. We regret that we could not include three more O serotypes as the targets of the PickPen-IMS because the antibodies were unavailable when we started this study. However, we have found the antibodies are available recently. The system can therefore be expanded to include the three additional antibodies so that the targets will cover the 15 important O serotypes in the future. We think addition of the antibodies rose to the three or more O antigens, if needed, are possible although the optimal conditions (i.e. bIMBV) and results of the detection method may change. Using a similar PickPen-IMS, our colleagues successfully prepared a set of IMB targeting 69 different K antigens at once and demonstrated its utility by detecting the target organisms from seafood [38]. Therefore, this method may be expanded to include three more O serotypes as a targets totaling to the fifteen O serotypes proposed in the introduction to cover important O157 and non-O157 serotypes in different parts of the world.
We are grateful to Kensuke Ozawa, Fumio Gondaira, and Akira Oshima for valuable comments and technical assistance; and to Yohko Takeda, Natthawan Sermwittayawong and Pharanai Sukhumungoon and their graduate students for technical assistance. This research was supported, in part, by Kakenhi Grant-in-Aid for Scientific Research (KAKENHI 24249038) and from the Japan Society for the Promotion of Sciences and a fund for Research on international cooperation in medical science, Research on global health issues, Health and Labour Science Research Grants, the Ministry of Health, Labor, and Welfare of Japan (H26-KOKUI-SITEI-001).