We investigated the incidence and antibiogram profile of Serratia marcescens among hospitalized individuals, hospital environments and halls of residence of Obafemi Awolowo University in Ile Ife, Osun state. These were with a view to provide key information at molecular level that are of epidemiological and therapeutic importance.
Two hundred and twenty samples from clinical and non-clinical sources were collected with ethical clearance approval (ERC/2018/09/02) from the hospital advisory committee. They were cultured on sorbitol Mcconkey agar infused with 200 U/ml of colistin for the selective isolation of Serratia species. Non-duplicate colony was picked from each cultures and characterized biochemically with the use of microbat 24E kit to identify various species isolated. The susceptibility profile of the isolates against selected antibiotics were studied by the Kirby-baeur disc-diffusion technique and screened for extended spectrum beta-lactamase (ESBL) and carbapenemase enzymes using double disc synergy test (DDST) and modified carbapenem inactivation assay (mCIM) respectively. Results were interpreted. They were also profiled for selected resistance genes through polymerase chain reaction (PCR).
Serratia marcescens showed great susceptibility to meropenem (100%), of loxacin (100%), ciprofloxacin (90.5%) and gentamicin (84.6%), and total (100%) resistance to ceftazidime, cefuroxime, ampicillin and augmentin. All the isolates (100%) were ESBL producers while 33.3% demonstrated carbapenemase production. The incidence of ampicillinase (AmpC), cefoxitin variant (FOX-1) ampicillinase, cefotaximase (CTX-M), sulphyhydryl variable (SHV), and temoniera (TEM) beta lactamase resistance genes among Serratia marcescens were 15.4%, 0.0%, 53.9%, 38.5% and 15.4% respectively. Equally, four (30.8%) and one (7.7%) isolates expressed co-carriage of two and three different resistance genes.
The study concluded that occurrence of multidrug resistant S. marcescens pose a public health threat in the study area considering their carbapenemase production and carriage of various resistance genes. The expression and co-carriage of resistance genes makes these isolates potential resistance-genes donor to other microorganisms.
Serratia marscescens, ESBL, Depressed mutants, Resistant genes, Carpenemase production
β-lactamases are hydrolytic enzymes that inactivate β-lactam antibiotics in the periplasmic membrane of Gram negative bacterial cell, effectively rendering it harmless to the cell [1].
The most well-known and studied β-lactamases, extended spectrum β-lactamases have emerged as a real threat to common antibiotic therapies. Aminoglycosides resistance is common among Serratia sp. [2], either due to enzymes that modify the drugs or efflux mediated that reduce the antimicrobials uptake by pathogens [3]. Methylases (RmtB, RmtC, RmtA, ArmA), phosphotidyltransferase (APH), nucleotidyltransferase (ANT) and Acetyltransferases (AAC family) are particularly effective in denaturing tobramycin, kanamycin, amikacin, gentamin, streptomycin and arbekacin [2].
Serratia sp. are usually susceptible to carbapenems, some S. marcescens strains harbor chromosomal carbapenemases [4], others acquire extended spectrum β-lactamases (ESBLs) through the plasmid. Beta-lactam antibiotics induces ampC gene expression among Serratia sp. by a complex mechanism that involves cell wall recycling [2]. Strains with induced ampC expression were regarded as depressed mutants [5]. Upregulation and increased expression of ampC β-lactamase among S. marcescens occur due to mutation in processes involved in recycling the bacterial cell wall [6]. Such mutants are termed depressed mutants and are markedly resistant to carbapenems and cefoxitin [5].
The first S. marcescens chromosomal carbapenemase regarded as SME - 1 was identified in 1982 in England [2] and has been described in sporadic infections in the United States of America [4]. GES-1 was found in S. marcescens isolated from 15 patients in an outbreak in a Dutch hospital from 2002 to 2003 [7]. GES-1 exhibit reduced carbapenemase activity and hydrolyzes penicillin and broad - spectrum cephalosporins [2]. Cai, et al. [5] reported an outbreak in China involving 21 strains of Klebsiella pneumoniae carbapenemase (KPC-2) expressing S. marcescens carried on plasmids. Isolates recovered in their study were the same clone and were either resistant or fairly susceptible to carbapenems. KPC are chromosomal carbapenemase of K. pneumoniae [2]. Also, Su, et al. [8] reported an outbreak in South Korea caused by meropenem - resistant S. marcescens in 2005 among 9 patients. The isolates lacked carbapenemase, and meropenem resistance was suggested to be a result of overproduction of chromosomal AmpC enzyme and to loss of outer membrane protein F (OmpF) [8].
Quinolones and fluoroquinolones target the bacterial DNA gyrase (gyrA) and topoisomerase IV (gyrB) that are critical for DNA replication and transcription [9]. Mahlen [2] indicated the efficacy of quinolones in treatment of S. marcescens infections with 95% and 100% sensitivity to ciprofloxacin and levofloxacin respectively among isolates recovered from their studies within 2008 to 2010. Later, reports showed increasing rate of S. marcescens resistance to these drugs [10], and the presence of chromosomal determinants [10], mutational concept [11] and scenarios of plasmids acquisition [3] responsible for quinolone resistance. Begic and Worobec [10] elucidated the presence of chromosomal resistance nodulation cell division (RND) family (SdeAB, SdeCDE and SdeXY) efflux pump among quinolone resistant S. marcescens. Kim, et al. [11] reported that quinolone resistance among tested strains of S. marcescens was result of amino acids substitution on isolates' DNA gyrase (dyrA). Plasmid mediated efflux pumps were also responsible for quinolone resistance among S. marcescens [3]. Quinolone resistance in Serratia marcescens may also occur due to presence of qnr genes. The qnr genes including qnrA, qnrB, qnrS, qnrC, and qnrD are found in plasmids and Smaqnr in chromosomes are repetitive pentapeptide proteins that block quinolones from acting upon their targets, they confer low quinolones resistance among S. marcescens.
Ambler, et al. proposed a different approach to classification of β-lactamases considering the molecular structure of the enzymes [12]. They observed two different molecular patterns after sequencing the amino acids in the enzymes, consequently classed them into serine and metallo beta-lactamases. This serves as the basis of classification by Paterson and Bonomo as shown in Table 1 [13]. They divided β-lactamases into four classes, with three being serine β-lactamases while one included metallo-β-lactamases group. The three serine β-lactamases are inactivated by clavulanic acid and other inhibitors, and are found in the class A penicillinases and class D oxacillinases. Metallo-β-lactamase and carbapenemase in
Table 1: Resistance genes among Serratia marcescens isolates. View Table 1
class B possess a zinc atom as their active site. This is important in their hydrolytic activity, although they are inactivated by EDTA [14]. The first ESBL was discovered in Germany in 1987, and since then, more than a thousand have been described.
Many different ESBLs have been discovered, with almost all having variants. The most important ones are the CTX-M, TEM, and SHV [15], with CTX-M being described as 'the most important development of the last two decades in the field of antibiotic resistance.' It is noteworthy to point out that almost all of the work done in the investigation of ESBL in Africa has been limited to Enterobacteriaceae.
The sulphydryl variable ESBL (known as SHV) is most frequently encountered in Enterobacteriaceae, and not in non-fermenting Gram negative bacilli [16]. One of the first ESBL to be discovered, it was found to be derived from the chromosomal β-lactamase of K. pneumoniae, having a global distribution [17]. It has however been reported in Serratia spp. additionally, in Pseudomonas aeruginosa and Aeromonas baumannii with clinical significance [1,18]. Almost wholly restricted to Enterobacteriaceae, TEM and CTX-M enzymes have also been reported in Serratia spp.
Biofilms are complex multicellular component formed by microorganisms connected by an extracellular matrix [19]. Biofilm formation in Serratia marcescens is coordinated by quorum sensing (QS) system. Labbate, et al. [20] reported thinner biofilm unable to form cell aggregates and chains among serrawetin (swr) deficient Serrati liquefaciensa. Serratia marcescens were controlled by lipopolysaccharide and flagella mechanisms. Mutants of S.marcescens with defective wecA and flhD (fliR) genes critical for lipopolysaccharides and flagella synthesis respectively were unable to induce silkworms cell death in their experiments [21].
This study provides information on resistant genes in the isolates of S. marscescens using polymerase chain reaction (PCR) with dual implication in epidemiological and therapeutic importance.
A total of 220 samples were collected from hospitalized patients of Obafemi Awolowo University Teaching Hospital and the hall environments of Obafemi Awolowo University within October 1 to December 30, 2018 with an approval (ERC/2018/09/02) by the medical advisory committee. Samples collected were urine from catheterized patients, swabs of surfaces, door handles, sinks and soap containers from selected wards. Swabbed samples of the university hall environments including bathroom tiles, and door handles were also collected following standard microbiological methods. Urine samples were collected in sterile universal bottle and transported to the laboratory. Each swabbed sample were transported in bijou bottles containing 5 ml of sterile tryptic soy broth. All samples were transferred to the laboratory for immediate cultures on freshly prepared sorbitol MacConkey agar infused with 200 U/ml of colistin.
All samples were inoculated by spread plate method on Sorbitol MacConkey Agar (SMAC; Oxoid CM0813; Hampshire, England) infused with Polymyxin B (colistin) (200 U/ml) with slight modification. Following manufacturer's recommendation, sorbitol MacConkey agar powder were dissolved in sterile distilled water (5.15 g/100 ml) in an air-tight conical flask. The mixture was homogenized and autoclaved at 121 °C for 15 minutes. The molten agar was allowed to cool to 45 °C prior to addition of dissolved colistin. Vials of Polymyxin B powder (Alvogen; New Jersey, USA) 500,000 U were dissolved in sterile distilled water and added to sterile, molten but cooled SMAC to make 200 U/ml concentration. Each conical flask was shaken to ensure even distribution of colistin in the molten agar, and poured into sterile petri dishes. The agar plates were allowed to gel, and dried in laboratory oven (45 °C) to remove condensed water vapors. Each sample was inoculated by spread plate method. The inoculated agar plates were incubated at 28 ± 2 °C for 24 hours to enhance pigmentation of Serratia sp. [22]. A colony of pink/red sorbitol/non- fermenting pigmented or non-pigmented colony were picked per plate. Pure colonies of each isolate were obtained by sub culturing each isolate on freshly prepared nutrient agar prior to biochemical characterization for identification.
Cultural, biochemical characterization and Identification of Serratia sp. were carried out following procedures. Standard procedures on Gram staining, motility test, oxidase test, gelatin hydrolytic test, DNAse and Casein Hydrolysis were conducted.
Further identification of isolates was done using Microbact 24E identification (Basingstoke, England) which is a standardized micro-substrate system for the identification of Enterobacteriaceae and common miscellaneous Gram-negative bacilli. Each kit of 24 miniature biochemical tests identifies GNB to species with proven degree of accuracy. Organism identification is based on pH change and substrate utilization. Each well was inoculated with an isolate suspension that reconstitutes the media. During incubation at 37 °C for 24 hours and 48 hours, metabolic reactions produced colour changes that were either spontaneous or revealed by the addition of reagents. The results were recorded on report forms and interpreted using the Microbact Identification Package. E. coli ATCC 25922 served as the control strain.
Two to three isolated colonies from the 18-24 hours cultures were emulsified in 5 mls of normal saline; it was mixed thoroughly to prepare a homogenous suspension.
The microbact test kit used was 24E. With the aid of sterile micro pipette and sterile tips 100 µl of the homogenous suspension were added to each test strips. The wells lysine, ornithine, hydrogen sulphide, containing the homogenous suspension were overlaid with mineral oil. The strips were incubated for 24 hours and 48 hours at 37 °C. The results were read after adding necessary reagents such as indole, VP and TDA.
This test was performed in well 7 (ONPG) after reading the ONPG reaction. One drop of Nitrate Reagent A and 1 drop of Nitrate Reagent B were added to the well.
The 24E strip was read after 24 hours and the reactions were evaluated as "positive" or "negative" by comparing to the colour chart and recording the results. The following reagents were added; Well 8 (Indole production), 2 drops of indole (Kovacs) reagent was added and was evaluated within 2 minutes of the addition of the reagents. Well 10 (Voges-Proskauer-reaction), one drop each of VPI and VPII reagent was added and evaluation was done 15-30 minutes after the addition of the reagents. Well 12 (Tryptophan deaminase), one drop of TDA reagent was added and the test was evaluated immediately after the addition of the reagent.
An octal coding system was adopted for Microbact. Each group of three reactions produces a single digit of the code. Each digit of the code is formed by adding the indices of the positive reactions. A total of eight digits from the eight groups form each code number (octal code). These codes were entered into the software package from microbact. Percentage probabilities were recorded along with isolate IDs.
The antibiotic sensitivity profile of each isolates against selected antibiotics was performed by the Kirby Bauer Disc diffusion technique as described by and interpreted following the Clinical Laboratory Standards Institut. The isolates were inoculated on Muller Hinton agar (Oxoid, USA). Overnight culture of each isolate was adjusted to 0.5 McFarland standard in 0.85% sterile normal saline. Antibiotic disks used in this study include: Ceftazidime; 30 μg, Cufuroxime; 30 μg, Gentamicin; 10 μg, Ciprofloxacin; 5 μg, Ofloxacin; 5 μg, and Amoxicillin/Clavulanate; 30 μg, Nitrofurantoin; 300 μg, Ampicillin; 10 μg, Aztreonam (10 μg) and Cabapenems: Imipenem; 10 μg, and Meropenem; 10 μg all supplied by Oxoid, USA. The diameters of zone of inhibitions of each antibiotics were measured and interpreted following the Clinical Laboratory Standards Institute interpretative charts. Results were recorded as sensitive 'S' and Resistant 'R' following CLSI recommendations.
Isolates resistant to at least one cephalosporin and/or carbapenems used were screened for production of extended spectrum beta lactamases (ESBLs) by the double-disk diffusion test as described by CLSI and the results were interpreted following the CLSI guidelines. A swab of bacterial culture (adjusted to 0.5 McFarland standard) were spread - plated on a Mueller - Hinton agar plate, one disk containing ceftazidime (30 μg), were placed on the plate. The other disk, containing amoxicillin/clavulanic acid (30 μg) was placed alongside the first disk (center-to-center distance of approximately 30 mm). The inoculated plates were allowed to air dry for 5 minutes and incubated at 37 °C for 24 hours. An expansion of the inhibitory zones between the two antibiotic disks was recorded as positive for ESBL production for a tested isolate.
Isolate(s) resistant to imipenem or meropenem were selected for this screening. They were screened for carbapenemase production by the modified Carbapenem Inactivation methods (mCIM) as described by CLSI. Briefly, each test isolate was cultured overnight on blood agar at 37 °C, a 1 μl loop full of the test isolate was emulsified in 2 ml of Tryptone soy broth. The mixture was mixed to homogenize, a meropenem (10 μg) disk was added into the mixture with a sterile forcep. The mixture was then incubated for 4 hours at 37 °C. On completion of the incubation, 0.5 McFarland of E. coli NCIB86 in nutrient broth. The E. coli NCIB86 was inoculated on MHA plate and allowed to dry for 10 minutes. The meropenem (10 μg) disk initially dropped in TSB-Isolate mixture was removed with a sterile inoculating loop and placed on the MHA (previously inoculated with meropenem sensitive E. coli NCIB86) plate. The plates were incubated at 37 °C for 24 hours. The cultures were measured and interpreted with reference to CLSI guidelines. Test isolates showing inhibition zones 6-15 mm or pinpoint colonies within 16-18 mm are positive for Carbapenemase production, while clear zones greater than 18 mm were regarded as negative for Carbapenemase production.
Bacterial DNA was extracted via the boiling method. The thirteen (13) multidrug resistant isolates were grown on Nutrient agar for 18 hours. A single colony growth was picked and emulsified in 0.1 ml of sterile water, mixed and boil for 10 minutes. This was to lyse the cells. The resulting cell lysate was centrifuged briefly (10 minutes at 15,000 rpm), and 3 μl of the supernatant was used as the DNA sample for the PCR reaction.
ESBL producing isolates were amplified using blaTEM, blaSHV, and blaCTX-M, AmpC and FOX genes. The amplification process was carried out under the following conditions: Initial denaturation at 94 °C for 3 minutes, followed by 35 cycle of denaturation at 94 °C for 45 seconds, annealing at 54 °C (AmpC and FOX), 52 °C (for TEM, CTX-M and SHV) for 1 minute and extension at 72 °C for 1 minutes, and a final extension at 72 °C for 3 minutes. Each PCR product was electrophoresed on 1.5% (w/v) agarose gel. Gel was stained with 5% (w/v) ethidium bromide and visualized under UV light.
Data obtained were analyzed and presented as frequencies and percentages. Data were compared with the use of the two tailed Fishers Exact Test with the SPSS statistical program (version 22). All reported p-values were two-sided and a p-value of less than or equal to 0.05 was considered to be statistically significant Table 2.
Table 2: Multidrug resistance among Serratia isolates. View Table 2
The Serratia species were isolated from the diverse range of specimens between October and December 2018. All 21 (100%) Serratia species were isolated from the environment and none (0%) from humans. The organisms recovered are Serratia marcescens 13 (61.9%), Serratia fonticola 1 (4.8%), Serratia rubidaea 1 (4.8%) and other Serratia sp. 6 (28.6%) (Figure 1).
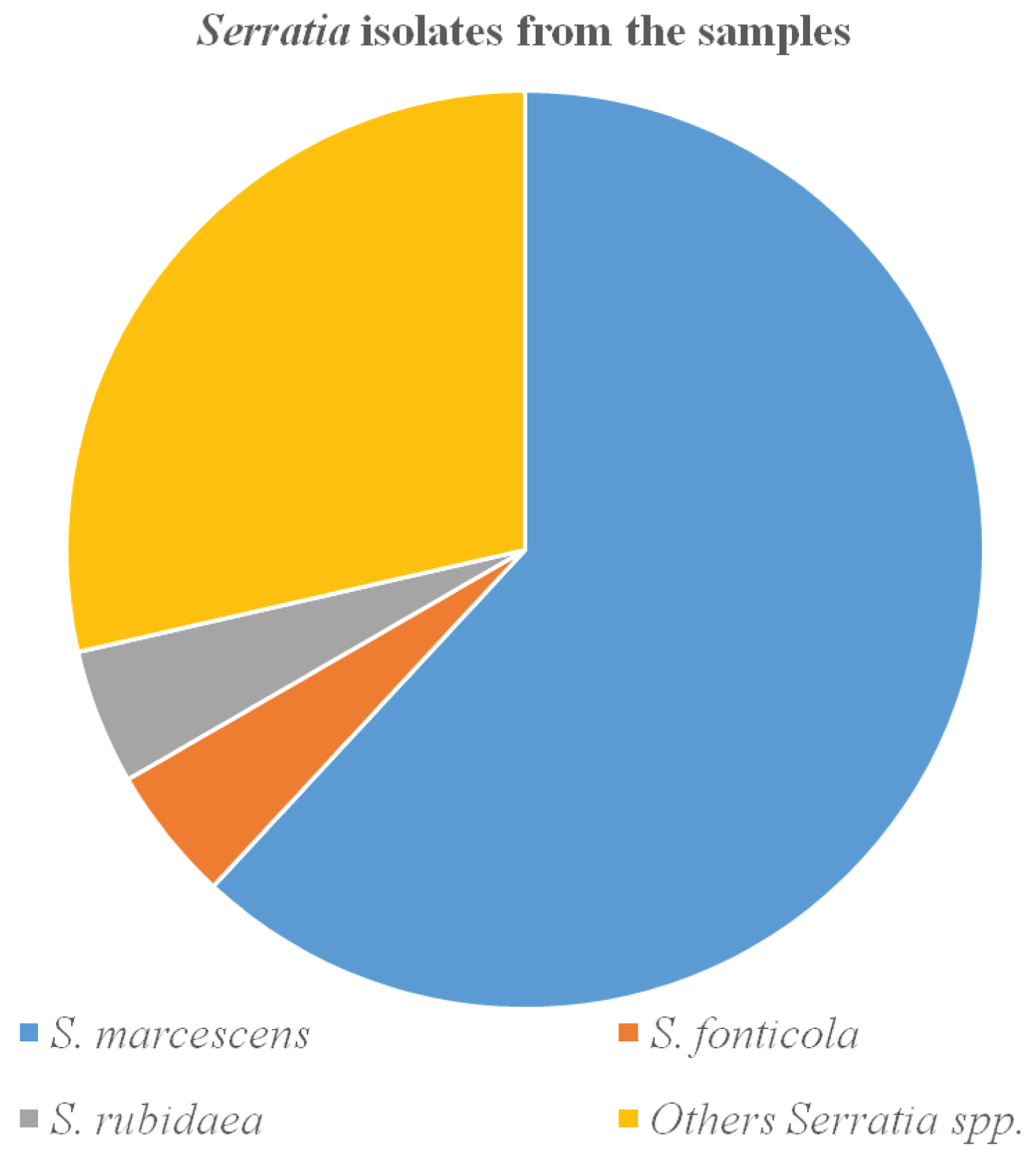 Figure 1: Distribution of Serratia species from the study area.
View Figure 1
Figure 1: Distribution of Serratia species from the study area.
View Figure 1
In this study, multidrug-resistant (MDR) status of Serratia isolates was tested against several antimicrobials. Accordingly, the overall rate of MDR among Serratia species was 90.48%. This means that 19.1% showed resistance to three antimicrobials, 38.1% showed resistance to 4 antibiotics and 9.5% of the Serratia sp. showed resistances to six antibiotics.
Phenotypically, all the 21 Serratia sp. were found to be ESBL producers after double disk synergy test (DDST) of which 2 (9.5%) were recovered from hospital environment and 19 (90.5%) were from environment of OAU community.
Seven (33.3%) of the isolates were demonstrated to be carbapenemase-producers with six (6) Serratia marcescens and one (1) Serratia fonticola.
All S. marcescens isolates were positive to ESBL production and 9 (69%) recovered showed carbapenemase production phenotypically.
Plates 1 to 3 shows the presence of various resistance genes among Serratia marcescens isolates. Five isolates showed the presence of two or more drug resistance genes. Serratia marcescens isolates designated as: PG4, MOZ2, DH5, I2.1 and ANG3 harbored at least two of the profiled genes (Plates 1-3). The incidence of TEM, CTX-M, SHV, AmpC and FOX genes among the isolates were 15.4%, 53.9%, 38.5%, 15.4% and 0.0 respectively as presented in Table 1 (Figure 2, Figure 3 and Figure 4).
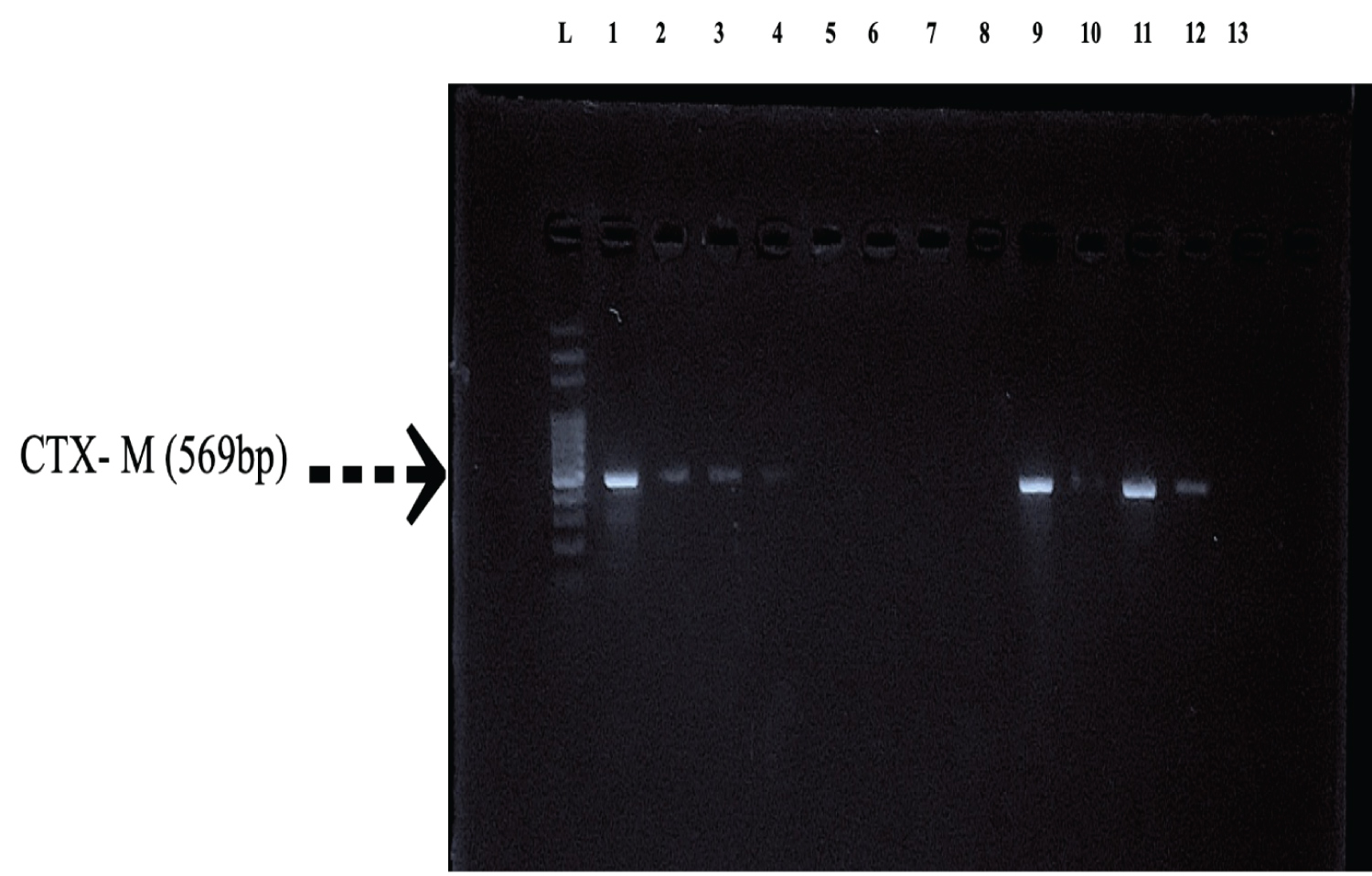 Figure 2: Characterization of ESBL enzymes (CTX-M) by PCR: Lane L: 100 bp ladder, Lane 1: DH5, Lane 2: E9, Lane 3: ANG3, Lane 4: PG4, Lane 5: MOR2, Lane 6: W5, Lane 7: MOZ4, Lane 8: W3, Lane 9: I2.1, Lane 10: MOZ2, Lane 11: B1, Lane 12: ANG2, Lane 13: F3.3.
View Figure 2
Figure 2: Characterization of ESBL enzymes (CTX-M) by PCR: Lane L: 100 bp ladder, Lane 1: DH5, Lane 2: E9, Lane 3: ANG3, Lane 4: PG4, Lane 5: MOR2, Lane 6: W5, Lane 7: MOZ4, Lane 8: W3, Lane 9: I2.1, Lane 10: MOZ2, Lane 11: B1, Lane 12: ANG2, Lane 13: F3.3.
View Figure 2
 Figure 3: Characterization of ESBL enzymes (TEM) by PCR: Lane L: 100 bp ladder, Lane 1: DH5, Lane 2: E9, Lane 3: ANG3, Lane 4: PG4, Lane 5: MOR2, Lane 6:W5, Lane 7: MOZ4, Lane 8: W3, Lane 9: I2.1, Lane 10: MOZ2, Lane 11: B1, Lane 12: ANG2, Lane 13: F3.3.
View Figure 3
Figure 3: Characterization of ESBL enzymes (TEM) by PCR: Lane L: 100 bp ladder, Lane 1: DH5, Lane 2: E9, Lane 3: ANG3, Lane 4: PG4, Lane 5: MOR2, Lane 6:W5, Lane 7: MOZ4, Lane 8: W3, Lane 9: I2.1, Lane 10: MOZ2, Lane 11: B1, Lane 12: ANG2, Lane 13: F3.3.
View Figure 3
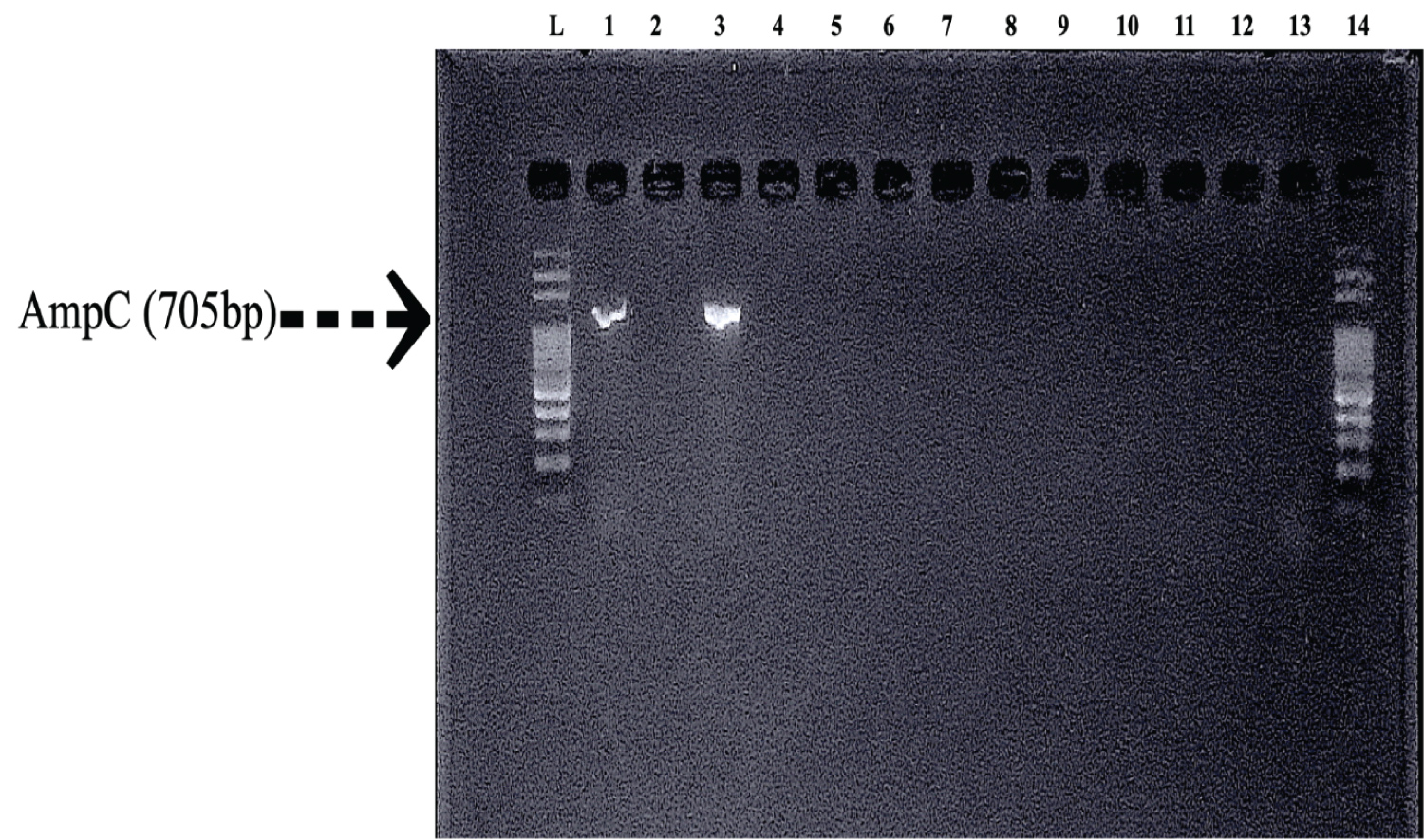 Figure 4: Characterization of AmpC Beta Lactamase enzymes by PCR: Lane L: 100 bp, Lane 1: DH5, Lane 2: E9, Lane 3: ANG3, Lane 4: PG4, Lane 5: MOR2, Lane 6: W5, Lane 7: MOZ4, Lane 8: W3, Lane 9: I2.1, Lane 10: MOZ2, Lane 11: B1, Lane 12: ANG2, Lane 13: F3.3, Lane 14: 100 bp ladder.
View Figure 4
Figure 4: Characterization of AmpC Beta Lactamase enzymes by PCR: Lane L: 100 bp, Lane 1: DH5, Lane 2: E9, Lane 3: ANG3, Lane 4: PG4, Lane 5: MOR2, Lane 6: W5, Lane 7: MOZ4, Lane 8: W3, Lane 9: I2.1, Lane 10: MOZ2, Lane 11: B1, Lane 12: ANG2, Lane 13: F3.3, Lane 14: 100 bp ladder.
View Figure 4
All (100%) the Serratia spp. identified and characterized in this study were from the environment and none (0%) from human unlike Dobretsov, et al. (2009) [23] that reported lower environmental occurrence.
All the isolated Serratia species in this study demonstrated very poor susceptibility patterns to most of the antimicrobials from major antibiotics classes tested using breakpoint Results from previous studies by Colodner, et al. [24] indicate that most ESBL producing Enterobacteriaceae are resistant to multiple antibiotic classes which was also observed in this study; where the Serratia isolates showed no susceptibility to the major classes of antibiotics; Penicillin (Ampicillin), Cephalosporin (Cefuroxime, Ceftazidime) and inhibitors of beta lactamase enzyme (clavulanic acid). This is a very alarming trend andpredicts that infections caused by S. marcescens may be difficult to treat.
There is variation in the susceptibility pattern of the Serratia species isolated in this study; S. marcescens were susceptible to meropenem, azithromycin, of loxacin, while the S. fonticola was susceptibility to of loxacin, gentamicin, nitrofurantoin, meropenem and azithromycin. This is in line report of Mahlen [2], where some strains of Serratia were sensitive to cefuroxime and resistant to Ceftazidime, some could be resistant to tobramycin and yet be resistant to gentamicin, this led Mahlen [2] to recommend that every Serratia infection treatment should be preceded by a laboratory sensitivity test. The overall MDR rate of Serratia species tested for seven classes of antimicrobial drugs was 90.5%. This finding goes in line to a study in Ethiopia [25] and Nepal [26] where 100% and 86.95% Enterobacteriaceae MDR rate were reported respectively.
Community-isolated ESBL - carrying strains are an emerging challenge for community medicine practitioners and hospitals. All the Serratia species isolated were found to be ESBL producer phenotypically. The spread of infections due to ESBL producers has been greater in countries with lower economic resources. This is very clear when comparing prevalence data from Sweden (3%) with those from Greece, Turkey, Portugal (> 25%) [13] or South America/West Africa (> 30%) [27,28]. Several reasons may account for this disparity: (i) Poorer social and economic conditions; (ii) Self-prescription of antibiotics, etc.
The worldwide dissemination of Enterobacteriaceae that produce ESBLs and specifically, CTX-M enzymes has been described as an unfolding pandemic [29,30]. In this study, 3 (Serratia marcescen) of the 13 isolates was found to have harbor BlaCTX-M. Enterobacteriaceae isolates expressing CTX-M beta-lactamases often display much higher resistance to cefuroxime against Ceftazidime [17]; in this collection of isolates, all of the cefuroxime-resistant Serratia marcescen were also resistant to ceftazidime. The prevalence of blaSHV and BlaTEM genes in this study was 19.0% and 9.5% respectively, which is a bit lower than the result obtained in the work done in Iran in 2010 with 26% and 24.5% prevalence respectively [31].
Multidrug resistance phenotype - producers were isolated from different Isolates, a testament to the ability of Serratia species to colonize the environments [22].
In the study done in a developing country (India) community carriage of Non fermentative Enterobacteriaceae was found more than the clinical which is similar to what was discovered in this study; as most (90.5%) the ESBL producing isolates were recovered from OAU (community) environment. All the ESBL producing isolates were recovered from OAU (community) environment.
Although expression of profiled ampC gene were observed from only 2 (15.38%) Serratia marcescens isolates from this study, they all showed phenotypic resistance pattern characteristic of ESCPM pathogens. This may indicate that isolates from this study have not undergone piperacillin/tazobactam induced mutations that results in depressed mutants characterized by high level of ampC expression.
Extended spectrum beta lactamase (ESBL) - producing Enterobacteriaceae is a growing risk for infection and resistance in the community in Ile-Ife. This study provides important data on Serratia spectra and antimicrobial resistance in a semi-urban setting in Osun state. Serratia species are important opportunistic pathogens; their multidrug resistance nature is well known and established, which has also been demonstrated in this study. Pan-drug resistance was also observed in some of the isolates. Enzyme production as a resistance mechanism was demonstrated in them, as they utilize it alongside other mechanisms to subvert the injurious action of antibiotics on them. Almost all antibiotics, except the tested fluoroquinolones and carbapenems were generally ineffective against Serratia marcescens in this study.
JOA Supervised and Coordinated all experiments and wrote the manuscript. ABO planned and performed the experiments. Both authors contributed to the development of manuscript.
We thank Dr. Folashade Osotimein of Albany State University, New York for her technical advice and assistance in procuring colistin used for the study.