Background: The association between genetic variants and respiratory conditions such as allergy and chronic rhinosinusitis has been the subject of extensive research. While the mitochondrial tRNA A3243G mutation has been extensively studied in the context of mitochondrial disorders, its potential role in inflammatory conditions such as allergy and chronic rhinosinusitis is not as well understood. In this study, we aim to explore and understand the potential link between the mitochondrial tRNA A3243G mutation and the prevalence of allergic CRS.
Methods: In the present case-control study, 214 allergic CRS and 238 healthy control subjects were included. Mitochondrial tRNA (Leu) A3243G mutation was investigated by PCR-RFLP method.
Results: The allergic CRS subjects showed association with other co morbid conditions asthma (10%), ear diseases (18%), hearing loss (12%) and diabetes (4%). However, the present study could not detect any mitochondrial A3243G mutation in subjects with allergic CRS and controls.
Conclusion: The findings indicate a lack of association between the mitochondrial tRNA A3243G mutation and the pathogenesis of allergic CRS. Further research is suggested to explore the functional analyses of mitochondrial gene mutations and their interactions with other genetic and environmental factors, particularly in the South Indian population.
Oxidative stress, mtDNA mutations, Allergy, Inflammation, Chronic rhinosinusitis
tRNA: transfer RNA; ATP: Adenosine Triphosphate; MELAS: Mitochondrial Encephalomyopathy Lactic Acidosis Stroke-like episodes; CRS: Chronic Rhinosinusitis; EP3OS: European Position Paper on Rhinosinusitis and Nasal Polyps; mtDNA: mitochondrial DNA; COPD: Chronic Obstructive Pulmonary Disease; OXPHOS: Oxidative Phosphorylation System; ELISA: Enzyme Linked Immunosorbent Assay; PCR-RFLP: Polymerase Chain Reaction - Restriction Fragment Length Polymorphism
Mitochondria are autonomous organelles that play a crucial role in various cell functions, including nutrient sensing, ATP production, calcium maintenance, stress response and are the major endogenous source of oxidative free radicals. They also regulate signaling pathways involved in cell fate determination, cell proliferation, cell differentiation, cell death.
Mitochondria is also tightly linked with inflammation, a complex biological response of the body to harmful stimuli, which can be part of many different diseases, including airway diseases, autoimmune disorders, metabolic syndrome, and cardiovascular diseases [1-3]. Human mitochondrial DNA (mtDNA) encodes for 13 essential polypeptides of the oxidative phosphorylation system (OXPHOS), as well as 2 ribosomal RNA (rRNAs) and 22 transfer RNA (tRNAs) for mitochondrial translation in the respiratory chain. The A3243G mutation is a well-known mutation that decreases the fraction of aminoacylated tRNA (Leu (UUR) which ultimately affects the entire respiratory chain and compromises the production of energy in the form of ATP. The mutation can also lead to various metabolic complications due to impaired oxidative phosphorylation, resulting in systemic effects on organs and tissues throughout the body. The mutation rate in mitochondrial tRNA genes is 10-20 times faster than in nucleic DNA due to increased exposure to free radicals, lack of protective histone proteins, and absence of introns except for the D loop. Over 150 different pathogenic mutations have been reported to be located within mt-tRNA genes [4,5]. Importantly, the mitochondrial tRNA A3243G mutation is typically inherited in a maternal fashion which is passed down from mothers to their offspring.
However, the extent to which individuals are affected can vary significantly, a phenomenon known as variable expressivity. This variation can be attributed to factors like heteroplasmy, which refers to the presence of both mutated and normal mitochondrial DNA within the same cell, tissue, or individual, resulting in a diverse range of clinical presentations and affected organ systems. The severity of the condition and the specific organs affected can depend on various factors such as the level of heteroplasmy and tissue distribution [5,6].
While the role of mitochondria and mitochondrial DNA (mtDNA) mutations in inflammation is an area of active research, the specific prevalence rates of the A3243G mutation in inflammatory conditions are not typically reported in the literature. Further, extensive research on in allergies and asthma showed involvement of numerous genes in the immune system signaling pathways and inflammatory pathways rather than mitochondrial genes It is also essential to note that inflammation and allergic conditions are influenced by a multitude of genetic and environmental factors [7,8]. Specific studies on the tRNA A3243G mutation in allergic and inflammation would contribute to a better understanding of the role mitochondrial genes in allergic and inflammatory respiratory disease such as chronic rhinosinusitis. To our knowledge, there is no report on the causal link between the tRNA A3243G mutation and allergic and inflammatory conditions like allergic chronic rhinosinusitis. Therefore, this study was carried out to elucidate the potential links between the mitochondrial tRNA A3243G mutation and allergic inflammatory disease such as allergic chronic rhinosinusitis.
To investigate the potential relationship between the tRNA A3243G mutation and allergic chronic rhinosinusitis, we conducted a comprehensive literature review of existing studies from PubMed, Scopus, and Web of Science examining the association of tRNA A3243G mutation in mitochondrial and inflammatory diseases. Molecular evaluation in the present case-control study has been performed in 214 adult allergic CRS subjects based upon the guidelines of European Position Paper on Rhinosinusitis and Nasal Polyps (EP3OS) and 228 healthy controls [9]. All the study subjects were diagnosed and confirmed by ENT specialists at MAA Hospitals Private Limited, Hyderabad. With high level of total IgE > 180 IU (ELISA) and with allergic symptoms such as nasal discharge, nasal itching and sneezing were defined as allergic CRS subjects. Subjects who showed normal total IgE level (< 180 IU) and with no symptoms of allergy and CRS were considered as controls. Cases with immunodeficiency diseases, immune-compromised conditions, nosocomial infections, COPD and allergic fungal sinusitis were excluded from the study. Informed written consent was obtained from all patients in accordance with the study protocol and the study was approved by the institutional ethics committee.
Approximately 2 ml of whole blood was used for DNA extraction using salting out method [10]. The present study has investigated mutation at mitochondrial tRNA (Leu) A3243G site involved in oxidative stress by using polymerase chain reaction (PCR) and polymerase chain reaction -restriction fragment length polymorphism (PCR-RFLP) methods. PCR was performed in a final volume equal to 25 μl of reaction mixture containing 1U of thermostable Taq DNA polymerase. To identify the mitochondrial tRNA (Leu) A3243G polymorphism, standard PCR was carried out using forward primer 5′-CAACTTAGTATTATACCCACAC-3′ and reverse primer 5′-ATTAGAATGGGTACAATGAGGA-3′. The amplification step of PCR was performed at 55 °C and after a total of 35 cycles, the PCR product of 162 bp was checked using 1% agarose gel electrophoresis. The PCR product was subjected for RFLP using restriction enzyme ApaI (Fermentas, United Kingdom). The presence of mtDNA A3243G mutation leads to the cleavage of the 161 bp PCR product into two fragments of sizes 87 bp and 74 bp respectively [11]. The restriction digest products were analyzed through 2-3% agarose gel electrophoresis containing ethidium bromide and visualized under ultraviolet light. Random PCR-RFLP retesting of approximately 10% of the samples per SNP was performed to find the concordance with the initial genotyping results. Two independent investigators who did not know whether the sample was obtained from the cases or control group scored the genotyping results. The data were compared between patients and controls using Microsoft excel windows 2010.
A total of 212 allergic CRS subjects recruited for the study included 130 male and 82 female subjects and age and sex matched 228 healthy controls (134 males and 94 females). The mean age of the allergic CRS subjects was 34.2 ± 12.10 years (18 to 65 years), and the mean age of the healthy subjects was 46.22 ± 12.16 years (18 to 81 years). The mean age of onset for allergic CRS subjects was 22 ± 10.28 years. The most common symptom observed in the allergic CRS subjects was nasal block (68%) followed by nasal discharge (64%), frequent cold (63%), Cough (21%), ear discharge (16%), ear block (12%), tympanic irritation (18%), and ear pain (10%), heaviness of head (11%), The co-morbid conditions asthma was seen in 10% of allergic CRS subjects while hearing loss was seen in 12% and diabetes was seen in 4% of the case subjects. There was no mitochondrial A3243G mutation detected in any subject with allergic CRS and controls. The demographic distribution of the allergic CRS subjects and controls is shown in Table 1 and associated CRS co-morbidities are shown in Figure 1. The agarose gel picture representing the results of PCR-RFLP is shown in Figure 2.
Table 1: Demographic and clinical characteristics of allergic chronic Rhinosinusitis subjects. View Table 1
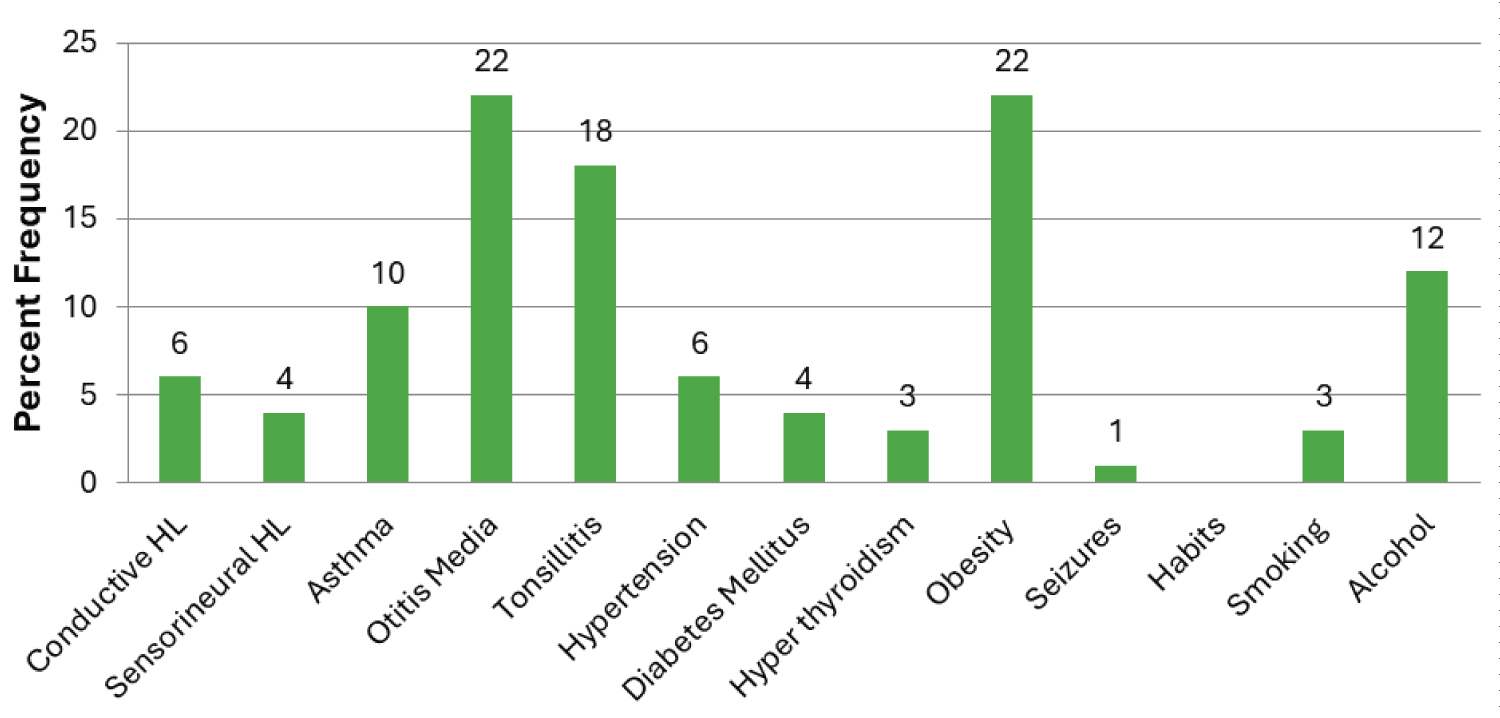 Figure 1: Distribution of comorbidities in allergic chronic rhinosinusitis subjects.
View Figure 1
Figure 1: Distribution of comorbidities in allergic chronic rhinosinusitis subjects.
View Figure 1
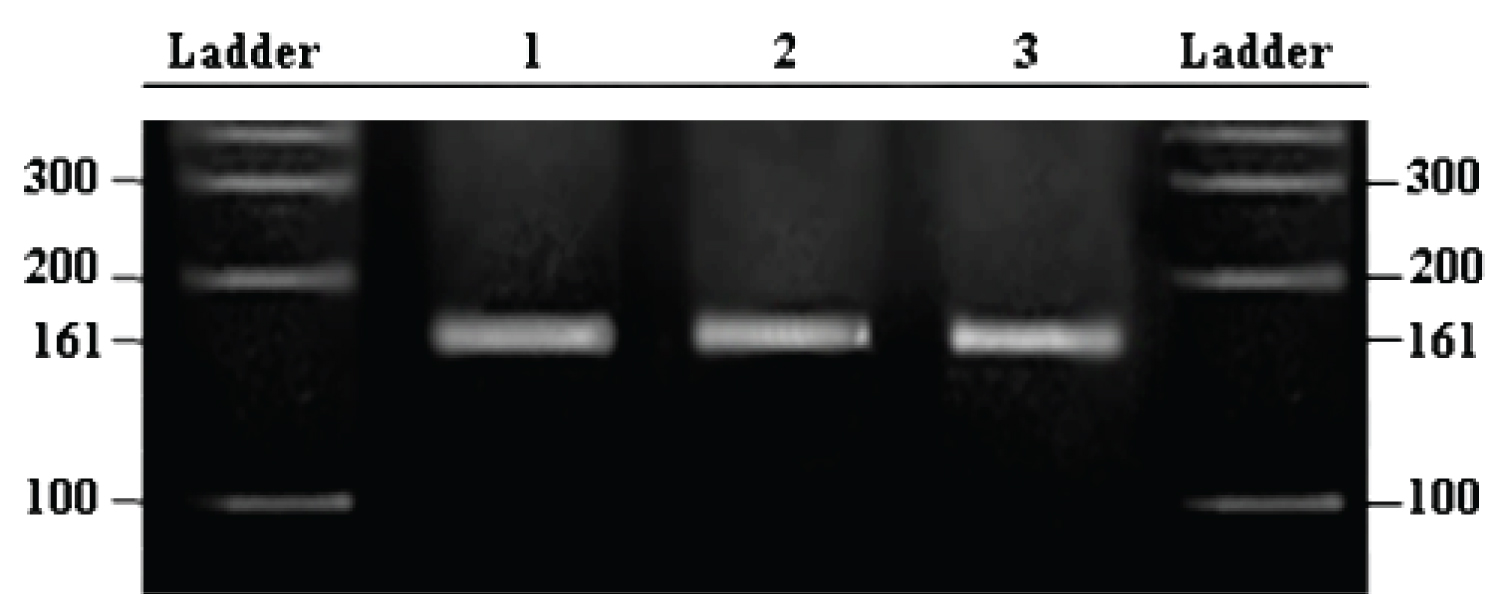 Figure 2: Agarose Gel picture showing PCR-RFLP products of mitochondrial tRNALeu (UUR) A3243G mutation digested by ApaI enzyme.
View Figure 2
Figure 2: Agarose Gel picture showing PCR-RFLP products of mitochondrial tRNALeu (UUR) A3243G mutation digested by ApaI enzyme.
View Figure 2
Mitochondrial tRNA Leu (UUR) A3243G mutation was reported to be strongly linked with high levels of reactive oxygen species (ROS) in patients with Mitochondrial Encephalomyopathy Lactic Acidosis Stroke-like episodes (MELAS) syndrome. The individuals carrying the mitochondrial tRNA A3243G mutation showed diverse range of symptoms of systemic disorders including mitochondrial myopathy, diabetes, and deafness due to endothelial dysfunction, pro-atherogenic and pro-inflammation [12-15]. Moreover, the mitochondrial tRNA A3243G and 1555(G) mutation was also shown to be particularly impactful on the nervous system leading to neurological symptoms such as seizures and sensory neural hearing loss respectively [16,17]. It's also worth noting that mitochondrial dysfunction could potentially lead to immune system dysregulation in asthma and allergic conditions where the body hyper reacts to harmless substances. Evidences suggest that ROS produced by various cell types as a consequence of their effector function, modulate innate and acquired immunity and thereby contribute to the pathogenesis of various airway diseases including asthma and allergic rhinitis [18,19]. Pollens and their extracts also generate ROS by their intrinsic nicotinamide adenine dinucleotide phosphate oxidases (NADPH oxidase) and induce antigen specific IgE production in allergic airway inflammation [20,21]. ROS can also regulate proinflammatory response produced by both upstream and downstream of Ca 2+ influx by pollen NADPH oxidase and the mitochondria, respectively [22]. Further, studies with metabolic inhibitors have demonstrated a close link between mitochondrial energy production and mast cell degranulation [23,24]. Exposure of the airway epithelium to particulates such as diesel exhaust particles induces ROS attributed to mitochondrial oxidative metabolism [25]. Further, protease-containing allergens in house dust mites, insects, pollen, pet dander can increase permeability and oxidative stress in epithelial layer of the nasal mucosa [26].
Mitochondrial A3243G tRNA Leu (UUR) lead to severe defect in respiratory chain assembly with abnormal incorporation of amino acids, alteration in stabilization, methylation, aminoacylation, and codon recognition of tRNA [20,27-29]. Strong influence of maternal history in development of asthma also raises the possibility that the mitochondrial dysfunction may contribute to the pathogenesis of asthma [30,31]. Moreover, evidence of association of mitochondrial haplogroup U with elevated total serum IgE levels in atopic phenotypes and in a cohort of children with mild to moderate asthma has been reported [31,32]. A study conducted by Mei, et al., (2017), identified the tRNA Thr G15927A mutation, tRNA Ala T5655C and tRNA Glu A14693G mutation were implicated to be associated with asthma in children of Guangdong Province, China [33]. However, Mkaouar-Rebai, et al., 2007, could not find association of any mutations in the mitochondrial tRNA (Leu (UUR) gene in the patients with mitochondrial diseases [34]. The present study also reports absence of association of mtA3243G mutations in allergic CRS subjects of South Indian population indicating further evaluation of mitochondrial mutations in allergic CRS. The pathophysiology of allergic CRS may be more influenced by environmental factors and other genetic components than by this particular mitochondrial mutation. Additionally, mitochondrial dysfunction may only play a role in a subset of patients with allergic CRS, and population differences may exist with mitochondrial tRNA A3243G mutations on different ethnic backgrounds in the pathogenesis of asthma and other diseases.
By investigating the relationship between this mutation and these respiratory conditions, we hope to contribute to the growing body of knowledge on the mitochondrial gene variants that may influence the development and progression of allergic CRS. The present study did not identify any association of mitochondrial tRNALeu (UUR) A3243G gene mutation with allergic CRS.
The author would like to thank ICMR for supporting in the form of ICMR SRF. The authors would also like to thank Mr. D. Dinesh and Ms. P. Padmavathi for their help in carrying out molecular work.
Raja Meghanadh Koralla: Conception of the work, diagnosis of the subjects, and critical revision of the article; Sunita G Kumar: Data collection and subject selection; Santoshi Kumari Manche: Help in conducting molecular work; Jujjuvarapu Venkata Ramakrishna: Help in conducting molecular work; Shehnaaz Sultana: Drafting the article; Madhavi Jangala: Conception of the work, drafting the article, Data interpretation; Pardhananda Reddy Penuguluru: Final approval of the version to be published.