Neonatal sepsis is a clinical syndrome that involves a systemic infection of the newborn during the first 28 days of life, with a high contribution of zoonotic sources worldwide. Sepsis is not, therefore, a monolithic condition. It encompasses a spectrum of illnesses that range from minor signs and symptoms to organ dysfunction and shock. Infection may occur by hematogenous, transplacental spread from a mother or by ascending infection from the cervix. Commonly, it is classified as early-onset (day of life 0-3) or late-onset (day of life 4 or later). The risk of acquiring the infection may be increased by maternal bacterial colonization, poor nutrition, exposure to animals, and poor hygiene.
Infection is a major cause of mortality during the first month of life and the mortality rate is still very high and extensively varies by setting. The prevalence of the infection is reported to have advanced in low- and middle-income countries more than in developed countries. In African countries like Egypt, Tanzania, South Africa, Nigeria, and Ethiopia, the prevalence of the disease has been remarkably reported to be higher.
The microbes that remain human competitors for dominance of the planet continue to cause neonatal sepsis. Globally, the most common isolated causative agents of neonatal sepsis were Escherichia coli, Staphylococcus aureus, and Klebsiella pneumonia, though Acinetobacter, Campylobacter jejuni, Shigella, and salmonella are also common. Given the diversified causes, less is known about host response to neonatal sepsis compared with other ages, particularly in relation to zoonotic diseases. However, immune system activation and inflammation are the major pathways leading to infectious and non-infectious stimuli. Regardless of the body response, establishing the diagnosis of neonatal sepsis using a clinical approach remains a commonly accepted practice. But different options are available to diagnose, implicate the present condition, and predict the prognosis.
Conclusion: Generally, while the need for an urgent commencement of antibiotics is one of the most pressing concerns in the control of an infection, a simple wash can be all that is needed to ensure your health is not compromised.
AMR: Antimicrobial Resistance; AP-PCR: Arbitrarily Primed Polymerase Chain Reaction; ATCC: American Type Culture Collection; BAC: Bacterial Artificial Chromosome; Bla: Beta Lactamase; CBC: Complete Blood Count; CLSI: Clinical Laboratort Standards Institute; CoNS: Coagulase-Negative Staphylococci; CRP: C-Reactive Protein; CTX-M: Cefotaximase Munich; EOS: Early-Onset of Sepsis; ESBL: Extended-Spectrum Beta-Lactamase; ESR: Erythrocyte Sedimentation Rate; FISH: Fluorescence In Situ Hybridization; GBS: Group B Streptococcus; JUMC: Jimma University Medical Center; LOS: Late-Onset of Sepsis; MDR: Multiple Drug Resistance; PCT: Procalcitonin; PCR: Polymerase Chain Reaction; PNA FISH: Peptide Nucleic Acid Fluorescence In Situ Hybridisation; UNICEF: United Nation Children’s Fund; WHO: World Health Organization; SHV: Sulfhydryl Variant; TEM: Temoniera; μg: Microgram
Neonatal sepsis is a clinical syndrome that involves a systemic infection of the newborn during the first 28 days of life. Whereas zoonosis refers to an infectious disease that has jumped from a non-human animal to humans and is common everywhere in the world. Regardless of the source, the syndrome of neonatal sepsis involves the physiologic alterations and clinical consequences of the presence of microorganisms or their toxins in the bloodstream or tissues [1-3]. The isolation of a pathogen from a typically sterile body fluid, such as blood or cerebrospinal fluid, has traditionally been used to define sepsis. The term systemic inflammatory response syndrome has been used to describe newborn sepsis since the clinical characteristics of sepsis can be caused by potent pro-inflammatory cytokines. Sepsis is not, therefore, a monolithic condition; however, encompasses a spectrum of illnesses that ranges from minor signs and symptoms to organ dysfunction and shock. It occurs in the interaction between invading micro-organism and the host immune, inflammatory, and coagulation responses. It has many aspects that contribute to epidemiological fragility, and it is described as the ultimate common death route of serious infectious diseases [4-6].
Commonly, neonatal sepsis is classified as early-onset (day of life 0-3) or late onset (day of life 4 or later). In newborns with early-onset sepsis, 85% of cases present within 24 hours, 5% present between 24 and 48 hours, and a smaller percentage of cases present between 48 and 72 hours [7]. The Early-onset sepsis is associated with mothers carrying microorganisms in their bodies that lead to infection in newborns. Infection may occur by hematogenous, transplacental spread from a mother or by ascending infection from the cervix [8,9]. Organisms that colonize the mother's genital tract may be acquired by the neonate as it passes through the colonized birth canal at delivery [10]. The risk of acquiring the infection may be increase by maternal Group B Streptococcus (GBS) colonization, poor hygiene especially after contact with young animals, climate change, prolonged rupture of membrane, premature birth, maternal urinary tract infection, chorioamnionitis, maternal fever greater than 38 °C (100.4 °F), low Apgar score (< 6 at 1 or 5 minutes), poor prenatal care, poor maternal nutrition, low socioeconomic status, low birth weight and congenital anomalies [7,11,12].
The late-onset of neonatal sepsis occurs within 4-28 days that some scholars consider to be 90-days-old (names it as late-late neonatal sepsis) of life and is often associated with the environment in which the neonate existed. The incidence of LOS is inversely related to the degree of maturity and varies geographically from 0.61% to 14.2% among hospitalized neonates [1,13]. Prematurity, central venous catheterization (duration > 10 days), nasal cannula or continuous positive airway pressure (CPAP) use, H2 -receptor blocker or proton pump inhibitor (PPI) use, and gastrointestinal tract pathology such as necrotizing enterocolitis (NEC) increases the risk of the infant for sepsis, having a catheter in a blood vessel for a long time, and staying in the hospital for an extended period of time predispose the neonates for the LOS. Moreover, toxins, whether proteinaceous or of a different molecular origin, play a crucial role in neonatal sepsis. Endotoxins from Gram-negative bacteria, such as lipopolysaccharide (LPS), activate monocytes and macrophages by binding to Toll-like receptor 4, whereas pore-forming proteinaceous exotoxins permeabilize target membranes of host cells, contributing to the sepsis phenotype. The phenotypes in neonatal sepsis exhibit a temporal specificity of gene expression that also known as stage specificity. The consequences of this cellular mechanism for cellular function and gene expression regulation are significant in epidemiology of neonatal sepsis especially for bacterial related sepsis [14-16].
With general overview, gene expression is different among different age groups where the neonatal and adult gene expression was more different than infant and child gene expression. Newborns showed a reduction in inflammation detection and signaling pathways compared to all other age groups. Study findings suggest that the transcriptomic response to sepsis is age-dependent and diagnostic and therapeutic efforts to identify and treat sepsis will have to consider age as an important variable. Dysregulated genes and the intensity of the transcriptomic response to sepsis from baseline indicate that the number of altered genes was proportional to age. As indicated in the Figure 1 -for instance the number of dysregulated genes in septic patients from baseline is proportional to age (Adults: 849, Children: 655, Infants: 513, Neonates: 226) [17,18].
 Figure 1: Proportion of gene expression in different age groups.
View Figure 1
Figure 1: Proportion of gene expression in different age groups.
View Figure 1
According to scientists, more than six out of every ten known infectious diseases in humans can be transmitted by animals, and three out of every four new or emerging infectious diseases in humans are transmitted by animals. Because their immune systems are still developing, neonates and young children are more likely to contract a serious illness from germs carried by animals. Despite the use of antibiotics and modern rehabilitation therapies and limiting contact with animals, sepsis is a major cause of illness (occurs in 0.5 to 8/100 live births) and death in neonates. However, the overall outcomes of sepsis has improved possibly due to greater focus on maternal intrapartum antimicrobial prophylaxis, early diagnosis using modern technologies and other improvements in supportive care [3,10,16,19]. But the mortality rate is still very high and extensively varies by setting. The prevalence of the infection reported to have more advances in the low- and middle-income countries than the developed countries. As reports from the different study showed, it ranked in the top 10 causes of mortality in the year 2014 with no significant change to date and imposed the highest-burden (4-10 times higher than the developed countries) morbidity in south Asia and sub-Saharan Africa in 2019. In Indonesia, about 5% of admitted cases were reported for neonatal sepsis with 28.3% of mortality among the cases [20]. The organisms most commonly associated with early-onset infection are Group B Streptococcus, Escherichia coli, Coagulase-negative Staphylococcus, Haemophilus influenza and Listeria monocytogenes. Epidemiological trends in early-onset sepsis indicate a reduction in the incidence of GBS disease, following widespread adoption of prenatal screening and treatment protocols [21,22].
In African countries like Egypt (45.9%), Tanzania (31.4%) [23], South Africa (LOS (86.8%) and EOS (13.2%)), and Nigeria (37.6%) the prevalence of the disease has been remarkably reported higher [24,25]. In a similar manner in Ethiopia, neonatal sepsis continued an important public threat across all regions whereof different time Hospital-based studies showed different prevalence for overall, early and late onsets of neonatal sepsis. Accordingly, the overall prevalence of neonatal sepsis was 63.69% (EOS comprised 59.33%) in 2017 [26] and 64.8% in 2019 [27] at the University of Gonder, 78.3% at Arba Minch General Hospital in 2018 [28], 33.8% in Wolayita Sodo in 2018 [29], and 77.9% in Shashamane (65% early and 35% late) in 2017 [30], and 52.6% at Jimma Medical center in 2019 (39.8% EOS and 12.8% LOS) [31]. In general, the epidemiology of the infection is influenced by at least four primary factors such as the pathogen, the pathogen burden, the infection site, and the host response. Etiologically, the late onset is concerned with Coagulase-negative Staphylococcus, Staphylococcus aureus, E coli, Klebsiella Pseudomonas, Enterobacter, Candida, GBS, Serratia, Acinetobacter and Anaerobes [32]. The infection trend from these group show an increase in coagulase-negative streptococcal sepsis, with most isolates showing susceptibility to first-generation cephalosporins. The source of infection for the late onset could be the infant’s skin, respiratory tract, conjunctivae, gastrointestinal tract, and umbilicus that become colonized via contact with the environment or caregivers. The microbes that remain human competitors for dominance of the planet continued causing neonatal sepsis. Of the competitors, bacteria, viruses, and fungi are involved in the causative lists of neonatal sepsis with varying degrees from region to region though polymicrobial sepsis is possible [33,34].
Bacterial etiologic agents, which include Gram-negative and, increasingly, Gram-positive organisms, are the most common causative agents of newborn sepsis in all regions. However, in the following table, the most common causes across the regions were detailed in Table 1 [3,12,19,24,35].
Table 1: Lists of bacteria commonly responsible for neonatal sepsis. View Table 1
In Bangladesh, a case-control study in 2020 showed Escherichia coli (40.7%), Staphylococcus Aureus (27.5%), and Klebsiella pneumoniae (18.7%) as the main isolated causative agents of neonatal sepsis [36] while Klebsiella pneumonia was the most common agent in India (48.1%) [37]. In Saudi Arabia, GBS was the commonest cause followed by E. coli [38] whereas in Egypt in 2012, Coagulase-negative staphylococci were predominant followed by Klebsiella pneumonia to cause neonatal sepsis [24]. While As a cross-sectional study report conducted in 2016 in Gonder-Ethiopia showed, Staphylococcus aureus (40%), coagulase-negative Staphylococci (21.6%), and Klebsiella pneumonia (15.8%) were commonly isolated etiologies of the neonatal sepsis [39]. In Arsi Hospital of Ethiopia, gram-positive bacteria were reported responsible for 53.4% neonatal sepsis with Coagulase-negative staphylococcus contributing 25% proven blood culture followed by E. coli (20.3%) and Staphylococcus Aureus (18.2%) [40]. Whereas in Jimma-Ethiopia, Klebsiella species (36.6%), Coagulase-negative, Staphylococcus (19.7%), and Staphylococcus aureus (18.3%) were reported the commonest isolate in 2018 [31].
From viral groups, a Lassa virus of zoonotic origin causes neonatal sepsis with 1% case-fatality rate while Herpes Simplex Virus (HSV) causes with common acquirement but less presentation during early neonatal life. Approximately 5% of neonatal HSV cases are acquired in utero, 85% are acquired peripartum, and 10% are acquired postnatally. Neonatal infection with enteroviruses and parechoviruses is not rare and has been reported to occur in infants within 29 days of age. Nowadays, several studies added the corona virus into the member of causes of neonatal sepsis. As the disease shed from asymptomatic individuals in 44% of cases neonates are likely to acquire the virus easily with unknown infector-infectee transmission pair [41-43].
Fungal EOS sepsis is an uncommon cause of neonatal sepsis. However, the presence of fungal colonized maternal genitalia during vaginal delivery and contaminants in the delivery settings by fungal infections could predispose the neonate. Candida species were dominantly recognized fungus in blood cultures and on blood agar plates and higher than 40% with most of them carrying risk of candidal meningitis [44,45].
Less is known about host response to neonatal sepsis compared with other ages for a number of reasons, particularly related to zoonotic causes while the main one being the very different definition of the disease [19,46]. However, immune system activation and inflammation are the major pathways leading to infectious and non-infectious stimuli. In the case of infectious pathogens such as bacterial pathogens, they encounter many different environments when infecting the host. They then developed a number of regulatory strategies to ensure their survival against changes in their growing environment [47]. As a result, pathogens rapidly change gene expression to survive in each of these environments. An important mechanism is transcriptional regulation, in which transcription factors bind to promoter sequences to recruit or block RNA polymerase to/from the target promoter [48]. Bacteria also coordinate gene expression at the post-transcriptional and post-translational levels. These mechanisms of gene regulation provide distinct advantages to bacterial fitness by enabling bacterial pathogens to rapidly alter gene expression. For example, immediately after a stimulus is sensed by a bacterium, post-transcriptional regulation enables fast responses to adjust the expression and/or activities of proteins. Moreover, post-transcriptional regulation enables bacterial pathogens to fine-tune gene expression by uncoupling transcription and translation [15]. This is beneficial as the functionally related genes may be organized within an operon and be cotranscribed. Post-transcriptional regulation is a very important mechanism to control gene expression in changing environments. In the past decade, a lot of interest has been directed toward the role of small RNAs (sRNAs) in bacterial post-transcriptional regulation through RNA-binding proteins (RBPs) also play an important role in the regulation of gene expression at this level [49]. It was noted that changes in gene expression in regulating the human host's physiologic response to a pathogen are a complex factor that regulates unique gene expression patterns in neonatal sepsis. As a result, gene expression profiles continue to provide novel mechanistic insights into functional pathways involved in sepsis pathophysiology [47,50]. Thus, sepsis has been documented to reflect an extremely complex host response to an invading pathogen. For example, after exposure to an acute infectious attack, an infant initiates an immune response to block the invasion of the pathogen. Initially, local and peripheral leukocyte-recognition receptors are activated by the presence of microbial products. Leukocyte activation leads to transcription of genomic DNA into mRNA, resulting in protein production (eg, IL, acute phase reactant). During this phenomenon proinflammatory genes such as tumor necrosis factor alpha (TNF-α), interleukin-2 (IL-2), interleukin-6 (IL-6), interleukin-7 (IL-7), interleukin-10 (IL-10), and interferon alpha-1 (IFNA1) will be initiated [51]. The presence of altered DNA sequences (eg, polymorphisms) and patterns of mRNA production can then be correlated with a clinical outcome such as mortality to gain insight into genetic influences on and involved pathways in this complex pathobiology [50,52]. In the complex pathway, LPS (endotoxin) is the main component of Gram-negative bacteria's cell wall, while exotoxins are from Gram-positive bacteria. The following describes the molecular mechanisms by which LPS induces gene activation and thus the expression of inflammatory mediators. LPS initially binds to the acute-phase LPS-binding protein (LBP) in the plasma, the level of which appears to rise in response to the insult. Bound LPS is subsequently delivered to the monocyte (and neutrophil) CD14 surface receptor. LPS then interacts with the transmembrane signal transduction receptor Toll-like receptor 4 (TLR4), which exists in complex with the accessory protein MD-2 [53,54]. TLR2 has been implicated as the receptor for Gram-positive exotoxin. This complexing and binding of LPS subsequently activates a number of intracellular signalling pathways, including the IkB kinase (IKK), nuclear factor kB (NFkB) pathway and three mitogen-activated protein kinase (MAPK) pathways [54]. These pathways phosphorylate and activate various transcription factors (see below), including NFkB/Rel proteins, activator protein 1 (AP-1) and nuclear factor-interleukin 6 (NF-IL-6), thereby allowing rapid gene induction and the expression of inflammatory mediators, including cytokines, chemokines, lipid mediators, inducible nitric oxide synthase (type II NOS), enzymes and adhesion molecules. Recently, a number of other mechanisms by which LPS induces gene transcription and inflammatory mediator expression have been described, including mechanisms operating TREM-1 (triggering receptor expressed on myeloid cells 1) [55], macrophage transmembrane potassium channels, and the intracellular cytoplasmic proteins Nod1 and Nod2 [56]. Additionally, sentinel cells (e.g. monocyte, macrophage) sense pathogens via PAMPs or DAMPs binding to PRRs. Pathogen recognition receptors (PRRs) include TLRs (Toll-like receptor), RLRs (Rig-1-like receptors), and NLRs (NOD-like receptors). Pathogen associated molecular patterns (PAMPs) include LPS (lipopolysaccharide), LTA (lipoteichoic acid), DNA, and RNA. Damage/Danger associated molecular patterns (DAMPs) can also be sensed through TLRs and include uric acid (UA), heat shock proteins (Hsp), and HMGB-1. Signaling occurs through a series of second messengers and results in transcription and translation of cytokines and chemokines that amplify the immune response as in displayed in Figure 2 [54,57].
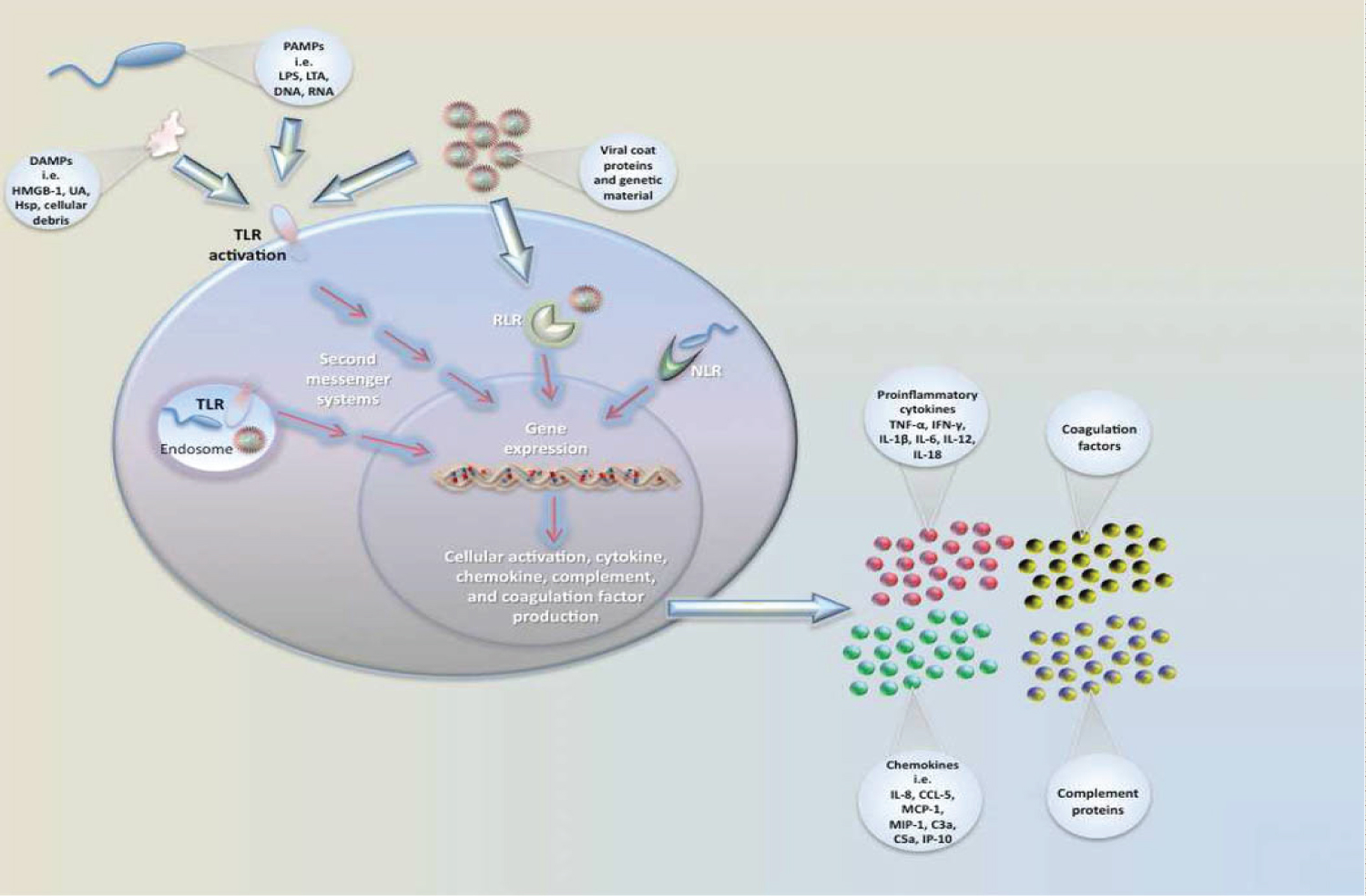 Figure 2: Neonatal immune responses to pathogenic insult and sentinel.
View Figure 2
Figure 2: Neonatal immune responses to pathogenic insult and sentinel.
View Figure 2
The clinical presentations are referred to as systemic inflammatory response syndromes. In the early stages, symptoms are often nonspecific (depend on the pathophysiology) and subtle, indistinguishable pathogens [5]. However, the symptoms of infection are demonstrated by the stage of the infection. The commonly demonstrated symptoms include but not limited to fever (presented in only 10-15%), diminished spontaneous activity, less vigorous sucking, apnea, bradycardia, temperature, instability, respiratory distress, vomiting, diarrhea, abdominal distention, agitation, seizures, and jaundice. Specifically, pneumonia is more common in early sepsis, while meningitis and bacteremia are more common in late sepsis. Early sepsis is 10 to 20 times more likely to occur in premature, low birth weight infants [33]. Most newborns will not experience long-term health problems if diagnosed and treated early for sepsis. However, if early signs or risk factors are missed, mortality increases. Neurologic damage to the brain or nervous system occurs in 15-30% of neonates with septic meningitis. Neonatal sepsis can kill up to 50% of infants who are not treated [57]. Infection is a major cause of mortality during the first month of life. It contributes to 13%-15% of all neonatal deaths. Low birth weight is associated with worse outcomes, as are gram-negative infections. Neonatal meningitis occurs in 2-4 cases per 10,000 live births and contributes significantly to mortality from neonatal sepsis; it is responsible for 4% of all neonatal deaths. Neurodevelopmental disorders are a concern in preterm infants with sepsis. Inflammatory molecules negatively affect brain development in patients of this age [58].
Laboratory-based diagnoses are the through for neonatal sepsis. However, because of the need for an urgent commencement of antibiotics, imperfect detection capacity, and related longer time of laboratory test, establishing the diagnosis of neonatal sepsis using a clinical approach remains a commonly accepted practice. Yet, different options are available to diagnose, implicate the present condition, and predict the prognosis of the sepsis [59,60].
History and physical examination is the cornerstone of clinical diagnosis for neonatal sepsis [37]. However, since some clinical manifestations in neonates are not a reliable indicator of illness from sepsis and the early signs of sepsis in newborns are non-specific, clinical diagnosis has often been ordered and treatments initiated before laboratory tests prove the presence of sepsis. In many settings, a precise clinical examination could be sufficient for diagnosis of neonatal sepsis and a positive test result would not dramatically increase the probability that the baby is infected [59,61]. In southeast Ethiopia, Arsi teaching Hospital, 34% of sepsis diagnoses were established clinically [40] while the approach was used in 96.08% of cases as indicated by a study conducted at Wollega teaching and referral Hospital [62].
To contribute to a progressive improvement of neonatal care, different laboratory diagnostic approaches are currently available, but the service varies based on the range and complexity of available tests that directly influenced by socio-economic status, technology advancement, available human skill, and institutional culture. The most common laboratory tests currently available are conventional microbiologic techniques, biomarkers from both neonatal and maternal side, Gene Expression Profiling, and molecular analysis [60,63]. A differential gene expression or DEGs can be valuable to pinpoint candidate biomarkers, therapeutic targets and gene signatures for diagnostics. Differential gene expression analysis revealed the genes involved in the immune response, chemokine-mediated signaling, neutrophil chemotaxis, and chemokine activity [17].
Diagnosis of neonatal sepsis continues to rely primarily on conventional microbiologic techniques from arterial or venous puncture can be used with good skin preparation to avoid cross-contamination. Total leucocyte count, absolute neutrophil count, and neutrophil ratio (immature-to-mature neutrophil ratio, I: M ratio, and immature-to-total neutrophil ratio, I:T ratio), platelet counts, and blood culture are commonly used tests. Of conventional microbiological methods, blood culture remains a definitive diagnostic tool for neonatal sepsis. However, the testing method is time-consuming (the 48-72 hours turnaround time) and produces false-positive results as well as false-negative results, which can be attributed to the difficulties in discriminating a true Coagulase-negative staphylococci (CoNS) infection from sample contamination [60,64]. The result of the infants can be false-negative in case of prior maternal antimicrobial exposure and inadequate amounts of punctured neonatal blood for test in about 75% of cases in infant who clinically fit the incidence of sepsis [60]. The incidence of culture-proven EOS in the United States was approximately 0.3-2 per 1000 live births [65]. In Ethiopian Hospitals like Arsi teaching Hospital (29.3%) [40] and Wollega teaching and referral Hospital only 3.92% the incidence of blood culture-proven test were limited [62].
Nearly 100% sensitivity and negative predictive value with high (> 85%) specificity and positive predictive value are the properties of ideal diagnostic biomarkers. Biomarker levels should change early in the disease course and remain altered for a period of time in addition to detecting the capacity of the etiologic types such as viruses, bacteria, or fungus. Biomarkers should also assist in the prediction of disease severity at the onset of infection and predict later prognosis with therapy with a quick turnaround time, small blood volume (< 0.5 ml), easily defined cut-off points, and ready availability with low cost. Wider studies revealed that none of a single marker significantly determines the diagnosis of neonatal sepsis over the other. Therefore, the use of the optimal set of biomarkers at a time increases the power of detection. From different existed tests Cytokines and chemokines (interleukin IL-6, IL-8, tumor necrosis factor-alpha), Cell Surface Markers (Cluster of Differentiation like CD14 & 64, and acute-phase reactants such as procalcitonin and C-reactive were the most practically used for diagnosis of neonatal sepsis [11,66-68]. From summarized data, about 50% of European and 90% of developing countries use different biomarkers for diagnosis of neonatal sepsis, while other studies also confirmed the presence of differences in determining the capacity of sepsis. Studies also identified that discrimination capacity of biomarkers was better in newer methods than the older methods, and of the biomarkers, CRP, PCT, and IL-6 were reported as the most commonly used tests globally as in Table 2 [67,69].
 Figure 3: Different molecular methods in the diagnosis of sepsis.
View Figure 3
Figure 3: Different molecular methods in the diagnosis of sepsis.
View Figure 3
Table 2: Diagnosistic methods and related biomarkers performance capacity for diagnoses of neonatla sepsis. View Table 2
The maternal serum has been tested as another potential non-invasive source for early detection of neonatal sepsis. Normal systemic inflammation of the maternal serum is associated with histologic chorioamnionitis, which is considered a risk factor for EOS. However, these serum inflammatory markers may increase after birth, reducing their usefulness in EOS diagnosis. A review and meta-analysis of inflammatory markers in maternal serum revealed that IL-6 in the cord blood sample alone was accurately compared and had a high positive likelihood ratio (5.47) in EOS cases [68].
Identification of sepsis through Gene expression profiling: Microarray-based gene expression is the prominent technique to culprit the etiologic agent and characterizes inflammatory signal pathways of sepsis [70]. Recently in medical practice, its high sensitivity (95%) and specificity (60%) guaranteed significance importance to reduce inappropriate antimicrobial usage. Literature indicate, Interleukin 1 receptor associated kinase 3 (IRAK3), angiotensin II receptor associated protein (AGTRAP), S100A9, Adrenomedullin (ADM), arachidonate 5-lopoxygenase (ALOX5), matrix metal peptidase 9 (MMP9) and ectonucleoside triphosphate diphosphohydrolase 1(ENTPD1) were reported as sepsis expressing genes [70].
Diagnostic molecular microbiology primarily focuses on the detection of fastidious and non-cultivable bacteria. Molecular pathogen detection methods predominantly evaluated in neonatal studies have used amplification methods such as PCR, rather than hybridization-based methods or mass spectrometry techniques. This method carries the promise of more rapid and sensitive results, particularly as molecular assays can be completed in less than 12 hours, and would be of utility in settings where there is pretreatment with antimicrobials, where low-density bacteremia occurs and culture-negative sepsis is common. Of the various amplification methods studied, broad-range conventional and real-time PCR have both been documented to provide a higher diagnostic sensitivity and specificity compared with other assays [11,71,72].
PCR, a technology based on the extraction of microbial DNA from blood samples and the subsequent sequencing or hybridisation of species-specific gene regions, is widely investigated for the detection of micro-organisms. Furthermore, real-time PCR which focuses on the temporal measurement of fluorescent signals generated in each amplification cycle, has been explored to monitor the microbial load and rapidly target specific micro-organisms in clinical specimens. Compared with the conventional culture technology, PCR technologies yield results with a higher sensitivity, a much smaller sample volume and less laboratory turnaround time. Recently developed PCR-based diagnostic platforms are highlighted by a low contamination rate, with DNA extraction, multiplex PCR and detection of PCR products performed in a closed system [33].
There is always something good to report when it comes to preventing and controlling zoonotic diseases. You can keep your kids healthy and still allow them to enjoy animals. The best way to protect yourself from getting sick from animals is to wash your hands thoroughly with soap and warm water after handling them, especially if you are handling young animals who are sick [3,73].
Neonatal sepsis involves a systemic infection of the newborn during the first 28 days of life, with zoonotic causes taking the lead worldwide. Infection may occur by hematogenous, transplacental spread from a mother or by ascending infection from the cervix. The risk of acquiring the infection may be increased by maternal Group B Streptococcus colonization and contact with animals, especially young animals with diarrhea. The mortality rate of the infection is still very high and extensively varies by setting. Globally, the most common isolated causative agents were Escherichia coli, Staphylococcus aureus, and Klebsiella pneumonia. Regardless of common Etiologic agents’ isolation, less is known about host response to neonatal sepsis compared with other ages. Immune system activation and inflammation are the major pathways leading to infectious and non-infectious stimuli. A simple wash can be all that is needed to ensure your health is not compromised.
The authors have no financial interest in any products mentioned or concepts discussed in this article.