A better understanding of the forces controlling cell growth will be essential for considering wound healing as a fundamental evolutionary with possibility of scar formation and reparative regeneration and the developing effective therapies in regenerative medicine and also in cancer. Historically, the literature has linked to cancer and tissue regeneration-proposing regeneration as both the source of cancer and a method to inhibit tumorigenesis.
Aim of this work is to verify similarity and difference between this processes an evolutionary approach. The same verify the evolution of some factors involved in cancer development. In all this process, genetically conserved or not. There are determinate kind of program (finalistic or afinalistic) whit a start messages but also a stop when the scope is achieved (regeneration).
It is clear that regeneration abilities in adult form is reduced in some superior vertebrates like humans and the same it seem related to an introduction of adaptative immunity.
This review discusses two powerful regeneration models, the vertebrate urodele amphibians and invertebrate, in light of cancer regulation.
Regeneration, Cancer, Stem cells, Wound healing, Reparative re-Generation, Invertebrates, Vertebrates, Pathology, Microenvironment, Genotypic-phenotypic expression, Heart regeneration, Re-expression embryonic markers, Diabetes mellitus
The term regeneration is a very interesting and interesting phenomenon in animals, which means a well-coordinated restoration of cells, tissues and organs that have been physically or functionally lost. This repair process should achieve the identification and recapitulation of the missing structures and at the same time achieve a functional integration between newly formed and already existing tissues in order to control physiological and structural changes.
In biology, evolution is the change in the inherited characteristics of a population from one generation to the next. These characteristics are the expression of the genes that are transcribed during reproduction and passed on to the offspring.
Mutations in these genes can create new or changing traits that lead to genetic differences (genetic variation) between organisms. New traits can also arise through the transfer of genes between populations, as in migration, or between species in horizontal gene transfer.
Evolution occurs when these genetic differences become more or less frequent in a population, either not randomly through natural selection or randomly through genetic drift. Natural selection is a process in which the genetic traits that contribute to survival and reproduction become more common and harmful traits become rarer. Over many generations, adjustments are made through a combination of successive, small, and random changes in characteristics, with the natural selection of variables best suited to their environment. In contrast, genetic drift results in random changes in the frequency of traits in a population. Genetic drift arises from the role that opportunity plays when a given individual survives and reproduces. The theory of evolution through natural selection was proposed almost simultaneously by Charles Darwin and Alfred Russel Wallace and elaborated in Darwin's 1859 book on the origin of species. With the advent of the increasing application of computer science to the life sciences along with the use of mathematical tools, computational models are also being developed to understand the process of regenerative decoration. Hence, there is a need for collaboration at the interface between life sciences, natural sciences and computer science to uncover the mechanism underlying the regeneration process.
In almost all animals it has been observed that aging is associated with a general decrease in tissue structure and function. This decline is believed to reflect a lack of selective pressure to preserve tissue beyond an age when the animal is likely to make a genetic contribution to future generations [1-3]. We also found that old animals significantly above reproductive age had little selective pressure to reduce the incidence of cancer [4]. For example, while mice can live 2 to 4 years in the laboratory and are prone to developing cancer in the second and third years, it is rare for a mouse to be more than a year-old in the wild [1].
Most wild rats die from other causes, such as starvation, the common cold, predators, or disease, before cancer becomes a possible cause of death in mice. Hence evolution favored the "early breed, often reproductive" strategy of the mice. Investing in better tissue management or preventing cancer well after a year has required the allocation of valuable energy early on, if that energy is best used for survival and reproduction in youth.
Cancer is caused by the process of somatic cell evolution as cell reproduction undergoes a series of genetic changes over time and spreads to create highly complex cancer [4,5]. Although many natural anti-cancer mechanisms have been developed [4], tumors have been reported in most metazoa [6]. Although there are some exceptional species, such as the naked mole rat and sharks, they have been claimed to be cancer resistant [7]. However, recent studies have shown that even these species can develop cancer [7,8], strongly suggesting that the vast majority of multicellular organisms are in fact prone to cancer.
Repeated recurrence of cancer in metazoa suggests that, like pathogens/parasites, the tumor can have a significant negative impact on the fitness of the host in the wild [9]. A recent review of wildlife cancer is supported by [10], which shows that the high prevalence of cancer in, for example, the Tasmanian Devils and Pelugas leads to an associated significant increase in death rate and decreased fitness. However, wildlife cancer statistics are very dispersed in the scientific literature and are therefore difficult to access.
Regeneration is a trait found in various classes of phyla, orders, and species in the animal kingdom [11]. Invertebrates such as planiformes, crustaceans, invertebrates, echinoderms and insects are known to have a strong global regeneration potential. However, the regeneration capacity can vary widely even in a certain order. Even in flatworms, which regenerate heavily and can regenerate large parts of the body including the head [12], species with a limited regenerative capacity have been found.
Regenerative capacity is also differentially distributed in vertebrates. Newts, axolotls and zebrafish are well-known for their abilities to replace entire limbs, fins and other body parts following amputation [13]. In contrast, regenerative capacity is restricted in many mammals (Figure 1).
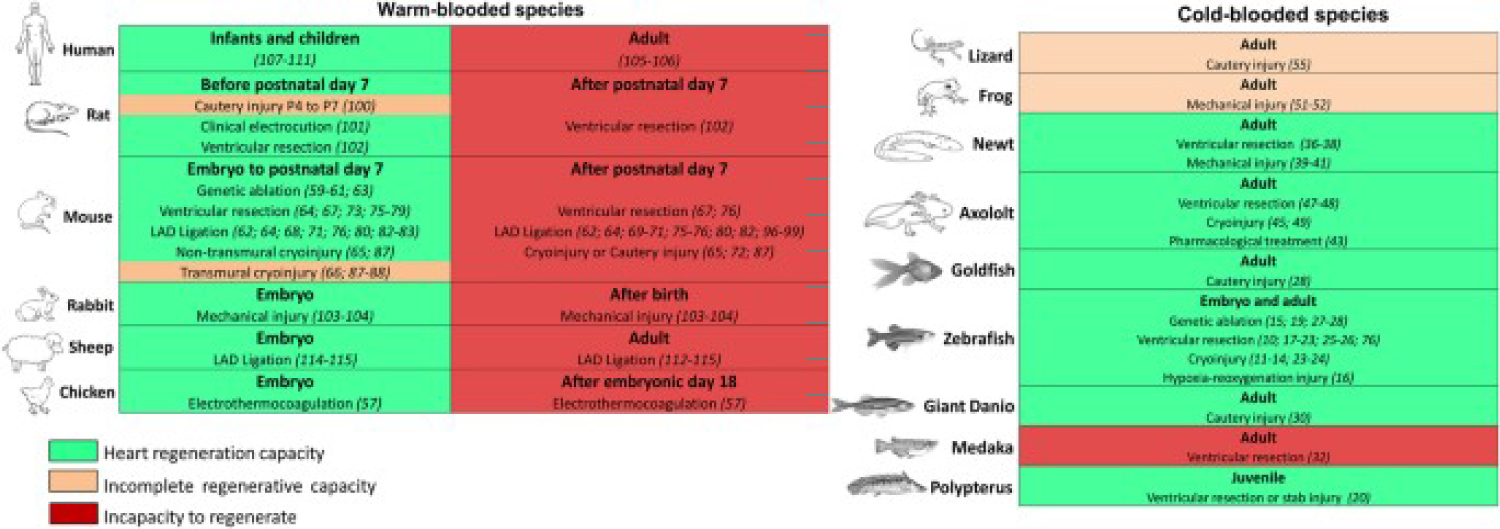 Figure 1: Warm- or cold-blooded animals are characterized by the ability to regenerate the heart. For each type, the heart's ability to regenerate is indicated by green (inability to regenerate), orange (inability to regenerate), or red (inability to regenerate). In each case, reference is made to the method of inducing cardiac injury and related references. In warm-blooded species, cardiac regeneration appears to be limited to a certain early growth period during fetal life and early neonatal life. In cold-blooded animals, six of the nine species can regenerate their hearts during puberty, while three of the nine species have incomplete or incapable cardiac regeneration [15].
View Figure 1
Figure 1: Warm- or cold-blooded animals are characterized by the ability to regenerate the heart. For each type, the heart's ability to regenerate is indicated by green (inability to regenerate), orange (inability to regenerate), or red (inability to regenerate). In each case, reference is made to the method of inducing cardiac injury and related references. In warm-blooded species, cardiac regeneration appears to be limited to a certain early growth period during fetal life and early neonatal life. In cold-blooded animals, six of the nine species can regenerate their hearts during puberty, while three of the nine species have incomplete or incapable cardiac regeneration [15].
View Figure 1
Adult mammals (including humans) are characterized by the regeneration of damaged tissues such as skeletal muscles and large parts of the liver, but have a very limited ability to regenerate some other organs. The heart is one of the least regenerative organs in adult mammals and there is a tremendous unmet medical need for cardiac regeneration treatments as cardiovascular disease remains the leading cause of death worldwide.
Most vertebrate species appear to have cardiac regenerative potential and are associated with decreased metabolic status, immature cardiac muscle structure, hypotension, immature immune system and hypoxia, and an inability to regulate body temperature. The relationship between these physiological parameters and the ability to regenerate is not well understood.
From an evolutionary point of view, it would be important to assess cardiac regeneration in a more diverse group of species, including birds, reptiles, and marsupials Inherent regeneration is critical in determining the ultimate repair effects in cardiac regeneration.
Different species have different cardiac regeneration abilities, and it is important to understand why animals such as salamanders and adult zebrafish have powerful cardiac regeneration skills in order to develop better treatments for patients with myocardial infarction and heart failure [14].
Comparisons of gene expression patterns between cancer and wound repair revealed important differences in many signaling pathways, from those associated with hypoxia-stimulating factor and insulin-like growth factor I to genes that determine morphology (e.g. CRYM, TCF21, CTGF, etc.) and glycolysis regulate sugar (for example PGK1 and HK1) [15]. For example, a list of candidate genes (such as cleft, snail, MITV, EDNRB, etc.) related to melanocyte development, regeneration, and cancer has been compiled [16]; However, all of these genes have yet to be characterized in large-scale regenerative models.
At Urodeles, it is believed that the application of modern molecular techniques will unlock the full potential of these model systems for studies related to carcinogenesis and regeneration. Accordingly, several candidate genes and signaling pathways for cancer have been identified in the planoids and molecular and genomic tools are currently in use [17]. Hence, the planoids are likely the first large-scale model of regeneration to study the molecular details of both regeneration and cancer in adults.
The highly desirable, still theoretical, cancer treatment is about the identification and destruction of abnormal cells without disturbing the balance. Indeed, leukemia cells can be specifically eliminated in mice with loss of PTEN function after rapamycin inhibition of the TOR (a vector component of the PI3K pathway) [18].
With an observational approach some relevant scientific literatures are analyzed to produce a global conclusion related the regenerative abilities of invertebrates and vertebrates useful in searching new therapeutic strategy.
Many tissue organs and apparatus are observed in different animal species and related their first phases of life (near birth) and next phases (adult or during ageing).
The same some similarity or difference of regeneration versus other process like neoplasm or wound scars process are analyzed.
Then finally is verified the inhibitory effect played by some tissue.
To control and stop the regeneration program to produce a physiologic functionally and anatomic replaces of a lost part or damaged.
Also the role played by introduction evolution of the adaptative immunity vs innate in regeneration abilities.
All literature was founded using PUBMED or other relevant biomedical database.
According to Baiping Cui, et al. [19]:
"For years, cardiomyocytes of postnatal mammals and humans were considered to be "terminally differentiated" and to be restrained in the G0 phase of the cell cycle throughout life. This assumption was changed several years ago by Bergmann, who applied 14C dating and proved the occurrence of cardiomyocyte renewal in the human heart, with a yearly rate gradually decreasing with age from 1% at 20 years of age to 0.4% at 75 years of age. Approximately 45% of cardiomyocytes undergo regeneration throughout life. However, the limited capacity of regeneration and proliferation of adult hearts still cannot compensate for the massive loss of cardiomyocytes in a single attack of MI. With the activation of repair-associated pathways following cardiac injury, the original injured sites of cardiac tissue are gradually occupied by fibrotic scars. In contrast to humans, zebrafish and salamanders, as vertebrates, possess a robust capacity of heart regeneration" [19].
"Inherent regeneration is critical to determine the final repair effects in heart regeneration. Different species possess various heart regeneration capacities, and understanding why animals such as adult zebrafish and salamanders have potent heart regeneration abilities is important to help us design better treatments for patients with MI and heart failure" [19].
According to Thomas P Lozito, et al. [20]:
As the closest relatives of mammals that exhibit enhanced regenerative-abilities as adults, lizards potentially represent the most relevant model for direct-comparison and subsequent improvement of mammalian healing.
Lizards are able to regenerate amputated tails, and exhibit adaptations that both limit tissue damage in response to injury and initiate coordinated regenerative responses. Reptiles and amphibians spontaneously regenerate cartilaginous skeletons in response to skeletal injury. The ability to regenerate whole appendages (limbs and tails) is a rarity among adult vertebrates. The most impressive examples of appendage re-generation are exhibited by the amphibians, including the urodeles (salamanders and newts) and anurans (frogs and toads). Salamanders and frogs are able to regenerate the limbs. Neotenic salamanders, which never fully develop and retain non-ossified, cartilaginous skeletons into adulthood, are able to regenerate fully formed limbs, with all the original cartilaginous skeletal elements of the originals.
Frogs, which do fully develop and exhibit ossified skeleton as adults, regenerate cartilage spikes rather than limbs following amputation.
Lizards are the only group of amniotes capable of tail re-generation as adults, and, unlike the amniotic salamanders, adult lizard axial skeletons are fully ossified. Both salamanders and lizards regenerate tails, and regenerated tail skeletons are almost completely cartilaginous. Salamanders regenerate cartilage rods ventral to regenerated spinal cords, while lizards regenerate cartilage tubes that enclose regenerated spinal cords. Regenerated tails of the less skeletally developed salamander segment and develop neural and hemal arches, and mature regenerated salamander tails are almost perfect copies of originals.
The more skeletally matured lizards, on the other hand, regrow imperfect regenerated tails, and lizard cartilage tubes never segment and are easily distinguishable from original tail skeletons. Non-mammalian vertebrate skeletal re-generation favors cartilage re-generation over bone. This is particularly interesting given that cartilage is a tissue that most mammals, and humans, are completely unable to heal, let alone regenerate.
Among the regenerative vertebrates, only lizards are grouped with mammals as amniotes, and that many of the regenerative properties and processes exemplified in lizards is shared with amphibians, the bulk of this review will focus on the lizard in its discussion of enhanced wound healing capabilities. Lizard tail re-generation follows waves of process of de-generation, proliferation, and differentiation [20].
According to Kathy Jacyniak, et al. [21]:
"Wound healing is an essential biological process involving the synchronized orchestration of numerous cellular and molecular events. While many of the key mechanisms involved in wound healing [including re-epithelialization (see Glossary), cell proliferation, angiogenesis, and extracellular matrix (ECM) deposition and remodeling] are widely conserved, the fidelity of repair often varies. For example, in humans and most other mammals, non-lethal injuries typically result in the replacement of damaged tissues with a fibrous substitute known as a scar. Although scars participate in re-establishing homeostasis and barrier functions, they lack the organization, tensile strength and specialized functions of the original. In contrast, other vertebrates-including various species of bony fish (teleosts), salamanders and lizards – are capable of wound healing without scar formation. Instead of replacing damaged tissue with a fibrous infill, these species undergo a tissue-specific program to restore tissue architecture and function. Although vertebrates lack the capacity for whole body regeneration, a broad range of organs can be partially replaced, including portions of the skin (epidermis and dermis), heart (ventricle), forebrain (telencephalon), spinal cord and even multi-tissue appendages, such as limbs and the tail Although it may be tempting to summarize scar-forming versus scar-free wound healing responses simply along phylogenetic lines (i.e. mammals scar, salamanders and lizards do not), the reality is far more complex. Fetal mammals can heal cutaneous wounds scar-free prior to the early- to mid-gestation period, while postnatal mice, rats, rhesus monkeys and human children can also spontaneously regenerate amputated digit tips (the distal phalanx: In addition, several species of African spiny mice are able to perfectly heal holes created in their ears, and even lose and then regenerate large portions of skin (60% of the total dorsal body surface area [21].
According to Hutchins ED, et al. [22]:
"Lizards, which are amniote vertebrates like humans, are able to lose and regenerate a functional tail. Understanding the molecular basis of this process would advance regenerative approaches in amniotes, including humans. We have carried out the first transcriptomic analysis of tail re-generation in a lizard, the green anole Anolis carolinensis, which revealed 326 differentially expressed genes activating multiple developmental and repair mechanisms. Genes involved in wound response, hormonal regulation, Musculo-skeletal development, and the Wnt and MAPK/FGF pathways were differentially expressed along the re-generating tail axis.
We identified 2 microRNA precursor families, 22 unclassified non-coding RNAs, and 3 novel protein-coding genes significantly enriched in the regenerating tail. High levels of progenitor/stem cell markers were not observed in any region of the regenerating tail.
We observed multiple tissue-type specific clusters of proliferating cells along the regenerating tail, not localized to the tail tip. These findings predict a different mechanism of re-generation in the lizard than the blastema model described in the salamander and the zebrafish, which are anamniote vertebrates. Lizard tail regrowth involves the activation of conserved developmental and wound response pathways, which are potential targets for regenerative medical therapies. Re-generation of appendages in the adult is observed in a number of vertebrates, including in the lizard tail, the salamander limb and tail, and the zebrafish caudal fin.
Molecular-cellular analyses in these model organisms are beginning to reveal conserved versus divergent mechanisms for tissue re-generation, which impacts the translation of these findings to the human therapies.
Re-generation in newts is associated with proteins specific to urodele amphibians, casting doubt on the conservation of these re-generative pathways with other vertebrates. Muscle formation during limb re-generation differs between newts and the axolotl.
Mammals possess some neonatal regenerative capabilities, including mouse and human digit tip re-generation and heart re-generation in the mouse, but these processes are limited in the adult-organism. Lizards are capable of regrowing appendages, and as amniote vertebrates, are evolutionarily more closely related to humans than other models of re-generation, (salamander, zebrafish).
An examination of the genetic regulation of re-generation in anamniote model will advance our understanding of the conserved processes of re-generation in vertebrates. In response to threats, lizards have evolved their ability to autotomize, or self-amputate, their tails and regenerate a replacement.
The patterning and final structure of the lizard tail is quite distinct between embryonic development and the process of re-generation.
Whereas the original tail skeleton and the muscular groups are segmentally-organized, reflecting embryonic patterning, the re-generated tail consists of a single un-segmented cartilaginous tube surrounded by un-segmented muscular bundles.
The segmental organization of the spinal cord and dorsal root ganglia in the original tail are absent in the replacement, with regenerated axon sex tending along the length of the endoskeleton.
De-differentiation has been proposed to be a major source of proliferating cells in the an amniote salamander blastema model. No clear evidence of de-differentiation has been identified in tail re-generation in the lizard, an amniote vertebrate.
A temporal-spatial gradient of tissue patterning and differentiation along the re-generating tail axis has been showed. While transcriptomic analysis has been carried out in anamniote regenerative models, including the zebrafish tail, the newt limb, and the axolotl limb, the genetic profile of pathways activated in re-generation of amniote appendages has not been well described.
Through transcriptomic analysis of lizard tail re-generation, it was identified that genes in pathways involved in developmental processes, myogenesis, chondrogenesis, and neurogenesis, adult processes, as wound and immune -responses, and are differentially expressed along the regenerating tail axis.
The Wnt pathway was significantly enriched along the regenerating lizard tail axis, and the activation of this pathway has also been verified in the salamander tail-tip and mouse digit tip re-generation. The activation of Wnt signaling in 2 amniote lineages, mammals and squamate reptilesm and urodele amphibians supports a role for this pathway in re-generation that is conserved among tetrapod vertebrates.
Transcriptomic analysis also showed that genes involved in thyroid hormone generation were differentially expressed, suggesting a regulatory connection between re-generation of the lizard tail and Musculoskeletal transformations during amphibian metamorphosis.
The lizarddio2 gene is the ortholog of deiodinase, iodothyronine, type I, which in mammals converts thyroxine pro-hormone (T4) to bioactive 3,3',5-triiodo-thyronine (T3). In Xenopus laevis, T3 is the key signal for the process of metamorphosis from tadpole to adult frog. Many of the changes associated with meta-morphosisare also observed in remodeling of the tail stump and outgrowth of the lizard tail. The lizard cga gene is the ortholog of chorionic gonadotropin, alpha chain, which encodes the alphachain of TSH and other crucial hormones.
During tadpole meta-morphosis, thyroid hormone (TH) and TSH rise, despite the normal expectation that TH would down-regulate TSH. Changes in TH regulation of TSH may also be altered in re-generation. Among the amniotes, the lizard retains genetic pathways associated with thyroid hormone regulation of meta-morphosis in amphibian vertebrates [22] (Figure 2).
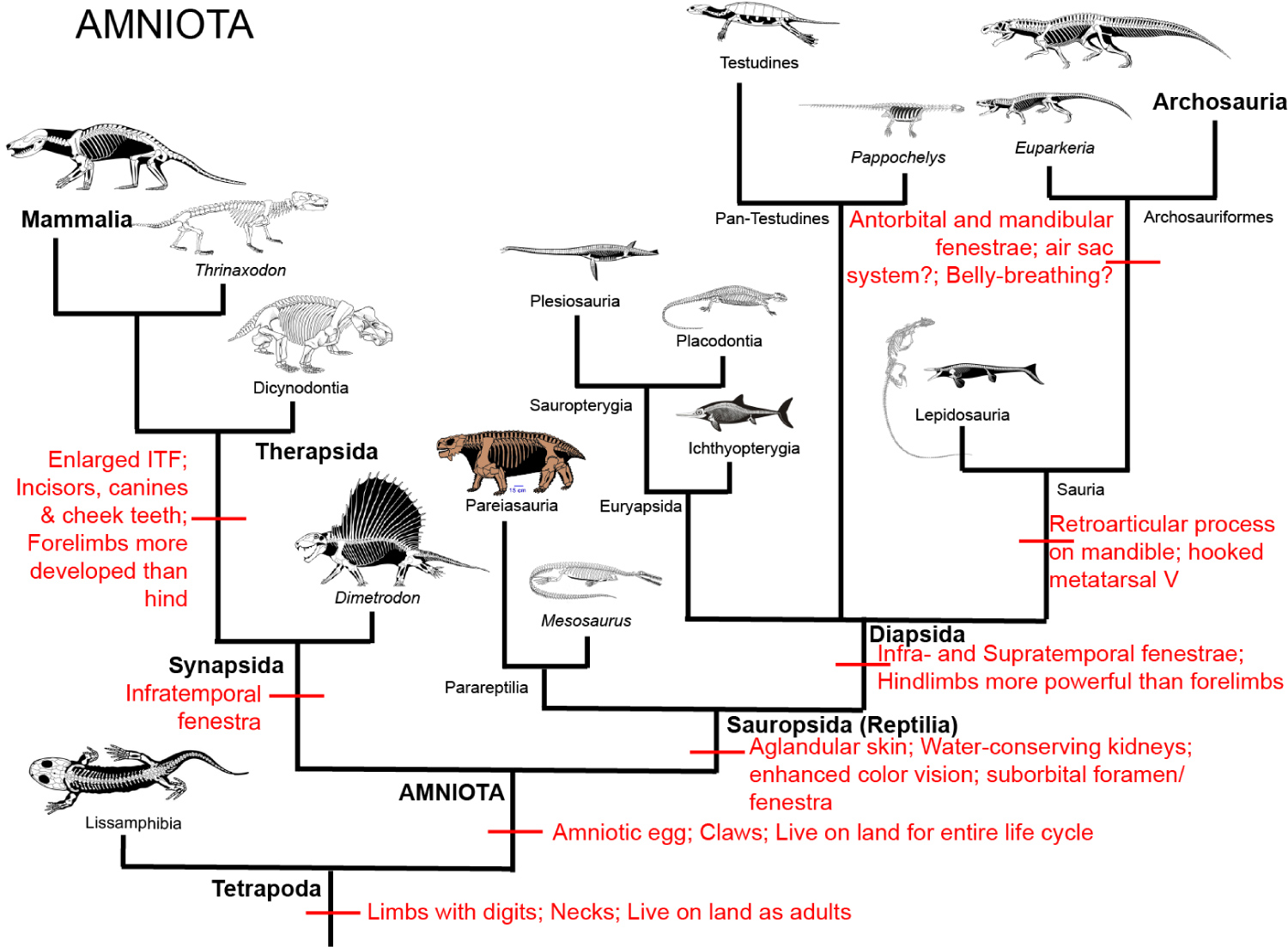 Figure 2: Amniota.
View Figure 2
Figure 2: Amniota.
View Figure 2
We previously identified conserved features in Notch pathway regulation of lizard and amphibian development, a gradient of these expressions in the pre-somitic mesoderm that was not observed in other amniote vertebrates and probably losted.
Transcriptomic analysis has showed activation of multiple genetic pathways, sharing genes that have been identified as regulating development or wound response processes in other vertebrate model. Some tissues are formed from patterning from a localized region of a single multipotent cell type, like the axial elongation of the trunk through production of somites from the pre-somitic mesoderm. Other tissues are formed from a distributed growth of distinct cell types, as development of the eye from neural crest, mesenchymal, and placodal ectodermal tissue.
The re-generation of the amphibian limb involves a region of highly proliferative cells adjacent to the wound epithelium, the blastema, with tissues differentiating as they grow more distant from the blastema. Re-generation of the lizard tail seems to follow a more distributed model.
Stem cell markers and PCNA and MCM2 positive cells are not highly elevated in any particular region of the regenerating tail, suggesting multiple foci of regenerative growth. This contrasts with PNCA and MCM2 immunostaining of developmental and regenerative growth zone models such as skin appendage formation, liver development, neuronal re-generation in the newt, and the regenerative blastema. Which all contain localized regions of proliferative growth.
Skeletal muscle and cartilage differentiation occurs along the length of the regenerating tail during outgrowth; it is not limited to the most proximal regions. The distal tip region of the regenerating tail is highly vascular, unlike a blastema, which is avascular. This suggests that the blastema model of anamniote limb re-generation does not reflect the regenerative process in tail re-generation of the lizard, an amniote vertebrate.
Re-generation requires a cellular source for tissue growth. Satellite cells, which reside along mature myofibers in adult skeletal muscle, have been studied extensively for their involvement in muscle growth and re-generation in mammals and other vertebrates. Re-generation of skeletal muscle in the axolotl limb involves recruitment of satellite cells from muscle. Mammalian satellite cells in vivo are limited to muscle, but in vitro with the addition of exogenous BMPs, they can be induced to differentiate into cartilage as well.
We have identified a coordinated program of re-generation in the green anole lizard that involves both recapitulation of multiple developmental processes and activation of latent wound repair mechanisms conserved among vertebrates. The process of tail re-generation in the lizard does not match the de-differentiation and blastema based model as described in the salamander and in zebrafish, but matches a model involving tissue specific re-generation through stem progenitor populations.
The pattern of cell proliferation and tissue formation in the lizard identifies a uniquely amniote vertebrate combination of multiple developmental and repair mechanisms [22].
According to Kazu Kik, et al. [23]
"CARDIAC REGENERATIVE CAPACITY IN VERTEBRATES
Mammalian hearts; In experimental settings. Adult mammals were probed for the capacity to regenerate cardiac- muscle after several models of injury, including MI, burning, freezing, mechanical injury, and chemical injury. Most researcher agree that this work and sub-sequent experiments to date involving modern capabilities to detect bona fide re-generation, generated little evidence to conclude that there is significant myocardial re-generation after cardiac injury. Most also agree that the key limitation to cardiac muscle re-generation is likely to be the poor ability of adult mammalian cardio-myocytes to enter the cell cycle and undergo division.
Cardiomyocytes in the fetal mammalian heart are mononucleated and proliferative; but shortly after birth the vast majority of cardiomyocyte DNA replication occurs without cytokinesis or karyokinesis. Most cardiomyocytes are binucleated with diploid nuclei in the adult mouse heart, and mononucleated with polyploid nuclei in the adult human heart. After this postnatal switch, it is rare for cardiomyocytes to enter the cell cycle. Observations suggest that injury may influence the propensity for adult mammalian cardiomyocyte proliferation.
In injured rodent ventricles, histological examination of 3H thymidine incorporation identified detectable DNA replication in nuclei of myofibers bordering necrotic tissue. Better resolution using transgenic mice in which cardiomyocytes were labeled by a nuclear-localized lacZ reporter protein, although no distinction between karyokinesis and cytokinesis was provided. These labeled cardiomyocytes were detected near the border zone of myocardial damage at exceptionally low levels (~ 0.0083%) [23].
According to Kurt Buchmann, et al. [24]:
Host responses against invading pathogens are basic physiological reactions of all living organisms. Even prokaryotes protect themselves by use of restriction enzymes and clustered regularly interspaced palindromic repeats (CRISPRs), being able to degrade invading foreign pathogens. Since the appearance of the first eukaryotic cells, a series of additional defense mechanisms have evolved in order to secure cellular integrity, homeostasis, and survival of the host. Unicellular amebae developed the ability to phagocytose foreign material as a part of their food uptake mechanisms and this basic phagocyte function is conserved in higher invertebrates and vertebrates in which the immunological function is more evident. Discrimination between self and non-self is also crucial for sexual functions securing genetic variation by exchange of genes between members of the same species. Recognition of non-self in both unicellular and multicellular organisms is based on cellular receptors allowing the host organism to bind, engulf, and/or kill potential invaders and offenders. Among the invertebrates, important groups such as protozoans (amebae, flagellates, and ciliates), sponges (such as bath sponges), cnidarians (e.g., jelly fish), worms (e.g., platy helminths, annelids, and nematodes), mollusks (snails and bivalves), crustaceans (e.g., crabs and prawns), chelicerates (spiders, mites), insects (e.g., flies), and echinoderms (seastars and seaurchins), are known to possess cells with receptors, which bind to foreign elements and allow differentiation of self and non-self. This ability is associated with presence of phagocytes bearing different names in various groups (amebocytes, hemocytes, coelomocytes, granulocytes, monocytes, macrophages), but basically they have a macrophage-like appearance and have, to a certain extent, comparable functions. Chordate evolution was based on the usage of existing genomes from ancestors and although deletions of significant parts of these have occurred, it is possible to trace some main lines from early and primitive organisms to highly developed mammals. The most primitive chordates comprising acranians (Amphioxus) and tunicates (ascidians) display a wide array of innate immune functions. In the primitive vertebrates comprising jawless fish, these functions became combined with an extensive use of leucine rich repeats (LRRs) as lymphocyte receptors. With the advent of cartilaginous and bony fish, the adaptive armament [major histocompatibility complex (MHC), immunoglobulins, T-cell receptors, extensive cytokine networks] appeared, and these new tools were further developed to a high level of sophistication through amphibians, reptiles, and birds to mammals [13]. This allowed a reduction of the copy number of many innate immune genes, but still the innate effector molecules have been taken into a complex network combining the obvious talents of fast acting ancient molecules with the highly developed specific recognition with memory seen in adaptive immunity [24].
According to Oviedo NJ, et al. [25]:
In his 1935 treatise, Waddington considers the mechanistic connection between uncontrolled cancerous growth and controlled embryonic development, postulating the existence of "individuation fields" that regulate tissue growth both during embryonic development and in adult tissues. Interestingly, Waddington's description of "individuation fields" is evocative of regenerative fields, and he himself links a field's strength to the organism's regenerative ability. Modern interpretation of Waddington's theory, which remains untested and largely overlooked in the current literature, implicates regeneration mechanisms as possible cancer regulators and underscores the need to investigate links between regeneration and cancer.
The term regeneration implies a well-coordinated restoration of cells, tissues, and organs that have been physically or functionally lost. This reparative process must accomplish the recognition and recapitulation of missing structures, while simultaneously achieving functional integration between recently formed and pre-existing tissues, in order to direct physiological and structural alterations. Furthermore, regeneration involving cellular proliferation (epimorphosis) requires instructive signals with the capacity to efficiently regulate cell cycle, resulting in a finite number of cells that undergo division and complete repairs. Participating cells must be precisely guided to needed areas, and once regeneration is complete specific cues are required to report regenerative success and signal termination. Otherwise, the initial response would continue indefinitely, causing undesirable consequences for body homeostasis. Diverse regenerative phenomena appear to utilize similar mechanistic procedures, including: Cellular replacement (e.g., physiological cell turnover), local tissue repair (e.g., epithelial wound repair), and regeneration of large sections (e.g., appendages and head).
Independent of magnitude, a regenerative event always seeks to maintain or reestablish both form and function (morphostasis). However, the process is not infallible, as demonstrated by growing evidence associating regeneration with cancer-related cellular abnormalities. Owing to space limitations, this review will be restricted to analyses of the relationship of abnormal cell proliferation and cancer to injury-induced epimorphic regeneration in adult animals [25].
Seemingly contradictory, regeneration might in fact both contribute to the source of abnormal growth and also provide a means to prevent and correct growth abnormalities. The initial phenomenological observations were summarized by Seilern-Aspang and Kratochwil, who surveyed the classical data and proposed two nonexclusive hypotheses: (i) The formation of malignant tumors derives from an impaired or incomplete regenerative process (Figure 3A), and (ii) The regeneration process may bring under control the autonomous growth of malignant cells (Figure 3B) [26]. The first hypothesis is largely based on observations of local tissue repair in mammals, where epithelial surfaces exposed to chronic damage or hypoxic conditions and inflammation result in growth aberrations during the regenerative response. It is important to note this is not universal to all cancers but is perhaps more likely in those of epithelial origin.
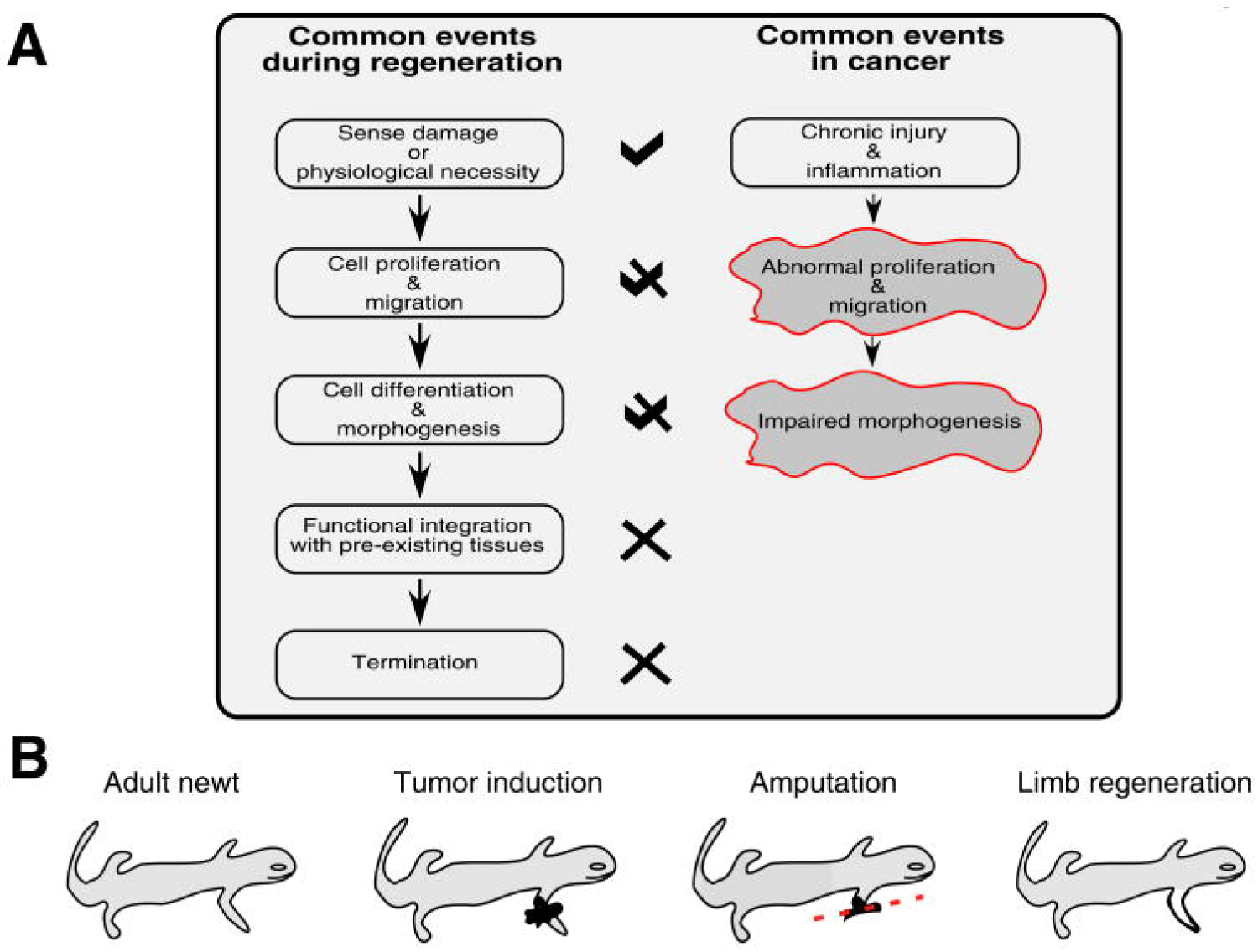 Figure 3: Links between regeneration and cancer [27] (A) Regenerative events and their corollaries in cancer. Importantly, the process of regeneration can be repeated without causing malignant transformation, while in cancer the regenerative process is incomplete such that chronic injury and inflammation leads to continuous proliferation. This suggests that characterizing signals at later stages of regeneration (especially those involved in termination) may help identify candidates able to stop abnormal proliferative responses to chronic injury; (B) Regeneration can correct malignant transformation, as in newt limbs where amputation through the site of induced tumors results in the regeneration of a normal limb without tumors [27] and references therein).
View Figure 3
Figure 3: Links between regeneration and cancer [27] (A) Regenerative events and their corollaries in cancer. Importantly, the process of regeneration can be repeated without causing malignant transformation, while in cancer the regenerative process is incomplete such that chronic injury and inflammation leads to continuous proliferation. This suggests that characterizing signals at later stages of regeneration (especially those involved in termination) may help identify candidates able to stop abnormal proliferative responses to chronic injury; (B) Regeneration can correct malignant transformation, as in newt limbs where amputation through the site of induced tumors results in the regeneration of a normal limb without tumors [27] and references therein).
View Figure 3
Comparisons of gene expression patterns between cancer and wound repair have revealed important differences in several pathways, from those associated with hypoxia-inducible factor and insulin-like growth factor-I, to genes regulating morphogenesis (e.g., CRYM, TCF21, CTGF, etc.) and glycolysis (e.g., PGK1 and HK1). For instance, a list of candidate genes (e.g. notch, slug, mitf, EDNRB, etc.) associated with melanocyte development, regeneration and cancer has been compiled; but all these genes remain to be characterized in large-scale regenerative models. In urodeles, modern molecular techniques need to be applied to release the full potential of these model systems for studies associated with carcinogenesis and regeneration. However, many candidate genes and signaling pathways associated with cancer have been identified in planarians, and the molecular and genomic tools are currently being applied [17]. Therefore planarians are potentially the first large-scale regenerative model for dissecting the molecular details of both regeneration and cancer in adults.
Recently, we discovered that using RNA interference (RNAi) to downregulate an evolutionarily conserved tumor suppressor gene in S. mediterranea, Smed-PTEN, leads to abnormal neoblast proliferation and lethal phenotypes. An important regulator of the PI3K signaling pathway, PTEN is among the most commonly mutated genes in human cancer. In planarians, loss of PTEN function is characterized by tissue disorganization, disruption of basement membrane integrity (alteration of epithelial-mesenchymal interactions), and the presence of abnormal cells that invade distant tissues to form aggressive and eventually lethal ectopic outgrowths. This phenotype is consistent with chemically-induced neoblastomas in planarians. In addition, neoblasts showed an impaired capacity to differentiate after Smed-PTEN RNAi [17], likely related to the abnormally invasive cells contributing to the epithelial dysplasias associated with advanced RNAi phenotypes (as well as the carcinogen-induced neoplasms). Just as with carcinogenic treatments, regeneration could be inhibited in animals subjected to Smed-PTEN RNAi, indicating that under certain conditions PTEN is also required for regeneration. A remarkable finding from these analyses was the striking regulatory conservation of the tumor suppressor function of planarian PTEN, which is similar to its mammalian counterpart but is absent in other invertebrates.
Controlling cell growth is clearly a central task in both cancer and epimorphic regeneration. Strangely, the regulatory mechanisms and strategies that animals have evolved to deal with cell growth regulation appear to have the capacity to either destroy or reconstruct. For instance, in mammals chronic epithelial injury often precedes malignant transformation, while in urodeles and planarians persistent damage generally ends merely with functional repair. Injury response always entails attempts to repair the damage, but the fundamental differences probably lay in the coordination of such responses rather than in structural or species-specific capabilities. In planarians, both carcinogenic chemical insults and manipulation of conserved cancer-associated signaling pathways have been shown to affect stem cell population behavior, leading to abnormal proliferation or even propagation failures.
According to Andreas Bergmann, et al. [27]
"Most metazoans have at least some ability to regenerate damaged cells and tissues, although the regenerative capacity varies depending on the species, organ, or developmental stage.
Cell replacement and re-generation occur in 2 contexts: Renewal of spent cells during tissue homeostasis (homeostatic growth), and in response to external injury, wounding, or amputation (epimorphic re-generation).
Model organisms that display remarkable regenerative capacity include amphibians, planarians, Hydra, and the vertebrate liver. Several mammalian organs including the skin, gut, kidney, muscle, and even the human nervous system have some ability to replace spent or damaged cells. Although the regenerative response is complex, it typically involves the induction of new cell proliferation through formation of a blastema, followed by cell specification, differentiation, and patterning. Stem cells and undifferentiated progenitor cells play an important role in both tissue homeostasis and tissue re-generation. Stem cells are typically quiescent or passing slowly through the cell cycle in adult tissues, but they can be activated in response to cell loss and wounding.
A series of works, mostly performed in Drosophila as well as in Hydra, Xenopus, and mouse, has revealed an unexpected role of apoptotic caspases in the production of mitogenic signals that stimulate the proliferation of stem and progenitor cells to aid in tissue re-generation [27]".
Mitogenic signaling by apoptotic cells may contribute to the formation of neoplastic tumors. The idea that tumors resemble wounds that do not heal is an old one, and many of the pathways involved in tissue re-generation and stem cell self-renewal, including Wnt and Hh, play prominent roles in human cancer. Wounding, inflammation, tissue stress, or tissue damage promotes apoptosis of cancer cells, these dying cells may release mitogens that promote malignant growth. The release of mitogens by apoptotic cells does not contribute to the overall growth of a tumor, it may stimulate proliferation of cancer stem cells or provide a supportive microenvironment for tumor growth. Virtually all cancer cells have acquired at least some degree of resistance toward apoptosis and hence may have features of so-called "undead" cells. Radiation therapy of cancer and chemotherapy, kill cancer cells by apoptosis and hence are expected to induce a "compensatory proliferation" response. This compensatory growth can explain oncogenic cooperation between genetically distinct cells in a Drosophila tumor model. The combination of apoptosis resistance and strong, therapy-induced cellular stress and damage may lead to the expansion of cancer stem cells and hence increase the likelihood of tumor re-generation and the formation of secondary tumors. Compensatory proliferation may play a role during different stages of carcinogenesis (including initiation, tumor promotion, and formation of secondary tumors) as well as in the response to cancer therapy, although the effects are likely to vary considerably in different paradigms.
Inflammation is commonly thought to promote tumorigenesis, and a complex relationship exists between inflammation and apoptosis. For example, hepatocellular carcinoma (HCC) frequently develops in response to chronic liver damage, and agents that induce cell death can promote HCC.
The chemical carcinogen DEN (diethylnitrosamine) induces hepatocyte death by DNA damage. DEN-induced hepatocyte death can activate the production of mitogens in adjacent myeloid cells, which in turn promotes compensatory proliferation of surviving hepatocytes.
Activation of both JNK and Stat3 has been implicated in several types of human cancer, and Stat3 function is critically required for the development of HCC. Because these pathways drive compensatory proliferation in Drosophila, and because caspase-3 and caspase-7 are required for liver re-generation in mouse, it is likely that caspases may regulate the production of some tumor-promoting cytokines.
Most animal cells have the ability to self-destruct by undergoing apoptosis, a morphologically distinct form of programmed cell death. The proper regulation of apoptosis is critical for both development and tissue homeostasis, and inhibition of apoptosis contributes to the development and progression of cancer. A central step for the execution of apoptosis is the activation of caspases, a family of cysteine proteases that are present as weakly active zymogens in virtually all cells. Caspases are activated by proteolytic cleavage of the zymogen in response to different stimuli, including developmental signals, as well as various forms of persistent cellular stress or injury, such as DNA damage, viral infection, hypoxia, increased presence of reactive oxygen species, loss of cellular adhesion, accumulation of unfolded proteins, excitotoxicity, shear stress, cytoskeletal damage, and other insults. Apoptosis can also be induced indirectly by cells that undergo necrosis in response to overwhelming physical injury. In all these cases, apoptosis appears to serve as an efficient cellular quality control mechanism that removes dysfunctional, unwanted, and potentially dangerous cells from the organism. Mammalian caspase is activated upon recruitment into the apoptosome complex with the adaptor protein Apaf-1 and cytochrome C.
An important event for apoptosome formation is the release of cytochrome c from mitochondria, a step that is regulated by Bcl-2 proteins. Once initiator caspases have been activated, they promote cleavage and activation of executioner caspases, such as caspase-3 and caspase-7, which then induce the demise of the cell.
Caspases are well conserved in the animal kingdom. In Drosophila, they are represented by one caspase-9 ortholog, termed Dronc, and 2 caspase-3-like proteins, Dcp-1 and DrICE. In addition to zymogen processing, an equally important layer of cell death control involves the inhibition of caspases. One important family of caspase inhibitors are the IAP (inhibitor of apoptosis proteins), which can bind to and inhibit caspases.
IAPs were discovered in insect viruses, but a family of related proteins was subsequently described in both insect and mammalian genomes. IAPs are characterized by the presence of at least one BIR (baculovirus inhibitory repeat) domain, which can directly bind to and inhibit caspases.
Several IAPs also carry a RING E3 ubiquitin ligase domain that promotes ubiquitination of key cell death regulators. In cells that are committed to die, IAPs are inhibited by a family of IAP antagonists: Reaper, Hid, and Grim, discovered in Drosophila.
Reaper, Hid, and Grim must localize to mitochondria to inhibit the apoptotic activity of IAPs. IAP antagonists contain an N-terminal IAP binding motif (IBM) that is necessary for binding to the BIR domains of and inhibiting Drosophila IAP1. Releasing the caspases Dronc and DrICE from Diap1 inhibition.
A similar IBM motif has been identified in mammalian IAP antagonists, including Smac and HtrA2 (caspase regulation is under dual control by both activating factors (Apaf-1 and cytochrome c) and inhibitory factors (IAPs), whose activity is in turn regulated by a complex ne2rk of upstream signaling pathways [27]".
According to Jonathan J Henry, et al. [28]
"Certain vertebrates are capable of regenerating parts of the eye, including the lens. Depending on the species, 2 principal forms of in vivo lens re-generation have been described wherein the new lens arises from either the pigmented epithelium of the dorsal iris or the cornea epithelium. These forms of lens re-generation are triggered by retinal factors present in the eye. Works have begun to illuminate the nature of the signals that support lens re-generation. This review describes evidence for the involvement of specific signaling pathways in lens re-generation, including the FGF, retinoic acid, TGF-beta, Wnt and hedgehog pathways." [28].
According to Chenxi Zheng, et al. [29]
"Bone and dental loss and defects are common and widespread morbidities, for which regenerative the rapy has shown great promise.
Mesenchymal stem cells, obtained from various sources and playing an essential role in organ development and postnatal repair, have exhibited enormous potential for regenerating bone and dental tissue. Mesenchymal stem cells (MSCs)-based bone and dental re-generation mainly include 2 strategies: The rescue or mobilization of endogenous MSCs and the application of exogenous MSCs in cytotherapy or tissue engineering. The diseased microenvironment not only impairs the regenerative potential of resident MSCs but also controls the therapeutic efficacy of exogenous MSCs, both as donors and recipients.
Approaches targeting a diseased microenvironment have been established, including improving the diseased niche to restore endogenous MSCs, enhancing MSC resistance to a diseased microenvironment and renormalizing the microenvironment to guarantee MSC-mediated therapies. The application of extracellular vesicles (EVs) as cell-free therapy has emerged as a promising therapeutic strategy. Over the past several years, MSC-based regeneration strategies have shown great promise for healing bone and dental loss and defects, both via endogenous restoration and exogenous transplantation.
Notably, the therapeutic efficacy of MSC-mediated regeneration is under tight control of the microenvironment, which not only regulates resident MSCs under both physical and pathological conditions but also modulates transplanted MSCs in cytotherapy and tissue engineering. As a result, achieving MSC-based bone and dental regeneration in diseased microenvironments remains a major challenge. Accordingly, microenvironment-targeting therapeutic strategies that may promote the optimization of MSC-based bone and dental healing in diseased microenvironments have been established. In this regard, several tactics have demonstrated enormous potential, including improvement of the endogenous microenvironment to revitalize innate MSCs, modification via pharmacological or epigenetic approaches to enhance exogenous MSC resistance, and restoration of the recipient microenvironment to benefit transplanted MSCs [29]".
According to Herrera SC, et al. [30]
"The JAK/STAT pathway is a conserved metazoan signaling system that transduces cues from extracellular cytokines into transcriptional changes in the nucleus. JAK/STAT signaling is best known for its roles in immunity.
In the early 1990s, several research groups demonstrated that a family of Janus (JAK) tyrosine kinases and a family of latent cytosolic transcription factors, termed signal transducers and activator of transcription (STATs), mediated interferon signaling in cultured mammalian cells. Subsequent work showed that numerous interleukins and growth factors also use JAKs and STATs to alter gene. Since these early studies, a wealth of research on various model organisms and cell types has provided further insight into how JAKs and STATs function to translate a multitude of signals into developmental or homeostatic responses. JAK/STAT signal transduction involves the binding of extracellular ligands to transmembrane cytokine receptors, which results in the activation of cytosolic JAKs and then of STATs. Activated STAT dimers translocate to the nucleus, bind specific DNA sequences in target genes and alter gene expression [30,31].
According to Rosenblad C, et al. [32]
"Both glial cell line-derived neurotrophic factor (GDNF) and its recently discovered congener, neurturin (NTN), have been shown to exert neuroprotective effects on lesioned nigral dopamine (DA) neurons when administered at the level of the substantia nigra. In the present study, we have explored the relative in vivo potency of these 2 neurotrophic factors using 2 alternative routes of administration, into the striatum or the lateral ventricle, which may be more relevant in a clinical setting. In rats subjected to an intrastriatal (IS) 6-hydroxydopamine (6-OHDA) lesion, GDNF and NTN were injected every third day for 3 weeks starting on the day after the 6-OHDA injection. GDNF provided almost complete (90-92%) protection of the lesioned nigral DA neurons after both IS and intracerebroventricular (ICV) administration. NTN, by contrast, was only partially effective after IS injection (72% sparing) and totally ineffective after ICV injection. Although the trophic factor injections protected the nigral neurons from lesion-induced cell death, the level of expression of the phenotypic marker, tyrosine hydroxylase (TH), was markedly reduced in the rescued cell bodies. The extent of 6-OHDA-induced DA denervation in the striatum was unaffected by both types of treatment; consistent with this observation, the high rate of amphetamine-induced turning seen in the lesioned control animals was unaltered by either GDNF or NTN treatment. In the GDNF-treated animals, and to a lesser extent also after IS NTN treatment, prominent axonal sprouting was observed within the globus pallidus, at the level where the lesioned nigrostriatal axons are known to end at the time of onset of the neurotrophic factor treatment. The results show that GDNF is highly effective as a neuroprotective and axon growth-stimulating agent in the IS 6-OHDA lesion model after both IS and ICV administration. The lower efficacy of NTN after IS, and particularly ICV, administration may be explained by the poor solubility and diffusion properties at neutral pH "[32].
According to Iaquinta MR, et al. [33]
"The regeneration of bone fractures, resulting from trauma, osteoporosis or tumors, is a major problem in our super-aging society. Bone regeneration is one of the main topics of concern in regenerative medicine. In recent years, stem cells have been employed in regenerative medicine with interesting results due to their self-renewal and differentiation capacity. Stem cells are able to secrete bioactive molecules and regulate the behavior of other cells in different host tissues. Bone regeneration process may improve effectively and rapidly when stem cells are used. Stem cells are often employed with biomaterials/scaffolds and growth factors to accelerate bone healing at the fracture site.
Osteoclastogenesis requires specific key mediators, such as the macrophage colony-stimulating factor (M-CSF or CSF-1) and the receptor activator of nuclear factor-kB ligand (RANKL or TNFSF11). M-CSF is produced by osteoblasts and many other cell types; it is necessary for the proliferation of osteoclast precursors, as well as their differentiation and fusion into osteoclasts.
RANKL binds its receptor RANK, which is localized on the surface of osteoclast precursors to allow fusion, maturation, survival, and osteoclasts activation. Osteocytes are the main source of the RANKL required for osteoclast formation. During bone resorption, several factors that lead to MSCs recruitment and differentiation are released through bone remodeling to enable bone formation in the bone marrow microenvironment.
The next transient phase, or reversal phase, consists in bone resorption inhibition in addition to osteoblasts recruitment and the subsequent differentiation that leads to bone formation. Osteoblasts can produce a protein called osteoprotegerin (OPG), which is a decoy receptor for RANKL. This protein prevents RANKL from binding to RANK, with the consequent inhibiting of osteoclast differentiation and activation. The final step in the remodeling cycle is represented by the termination phase, when an equal amount of resorbed bone has been replaced. Osteocytes contribute to ending the remodeling process by producing sclerostin, which inhibits the bone formation induced by Wnt signaling in osteoblasts. At the end of the process, mature osteoblasts undergo apoptosis, become bone lining cells or differentiate into osteocytes [33].
According to Guo-Ping Xu, et al. [34]
"In 1991, Caplan introduced the term "mesenchymal stem cells" to define the putative stem cells of skeletal tissues (bone and cartilage). The concept of MSCs extended to include bone marrow, adipose tissue, the periosteum, the synovial lining, muscle tissue, the umbilical cord, and different types of dental tissues. BMMSCs show an essential role in supporting bone healing through the secretion of nutritional and immunomodulatory factors rather than via a direct effect on the formation of the bone callus. BMMSCs secrete growth factors and cytokines to influence bone regeneration via paracrine and autocrine systems; this process includes vascular endothelial cell growth factors, platelet-derived growth factors, BMPs, fibroblast growth factors, insulin-like growth factor, and epidermal growth factor.
Inflammation is essential for any wound healing including bone repair. The first phase of fracture repair is the inflammation phase. Besides the trophic role, BMMSCs are critical regulators of the local inflammation micro-environment during bone repair. Macrophages are a key cell population that contributes to the inflammatory environment, whereas BMMSCs show an immunomodulatory effect on macrophages. These inflammation factors include prostaglandin-E2, monocyte chemoattractant proteins (MCP-1 and MCP-3), tumor necrosis factor-α, transforming growth factor-β, and numerous interleukins (IL-1, IL-3, IL-4, IL-6, and IL-10) [34-38].
According to Yunhao Qin, et al. [39]
"Stimulating bone growth and regeneration, especially in patients with delayed union or non-union of bone, is a challenge for orthopaedic surgeons. Treatments employed for bone regeneration are based on the use of cells, biomaterials and factors. Among these therapies, cell treatment with mesenchymal stem cells (MSCs) has a number of advantages as MSCs: 1) Are multipotent cells that can migrate to sites of injury; 2) Are capable of suppressing the local immune response; and 3) Are available in large quantities from the patients themselves. MSCs inhibit the immune response in two ways: by the contact immune response and by the non-contact immune response [39].
The rate of apoptosis of lymphocytes in an experimental group in which interleukin 6 antibodies were added to the co-culture environment was higher than in the control group treated with MSCs only. This result showed that, by neutralising IL-6, the immune inhibition effect of MSCs was weakened. The contact immune response stimulated the expression and secretion of IL-6 in MSCs, thus inhibiting lymphocyte apoptosis [39]."
According to Lopes D, et al. [40]
"Expression of the fibroblast growth factor (FGF) receptor 3 and a membrane-spanning tyrosine kinase receptor by chondrocytes. These cells contain a domain that binds to extracellular ligands, including FGFs, initiating the receptor's autophosphorylation, as well as the stimulation of the tyrosine kinase activity, leading to the inhibition of proliferation and growth of chondrocytes ; Indian hedgehog homolog (Ihh), a protein, present in the embryogenic patterning, controls the endochondral bone formation by inhibiting the differentiation of hypertrophic chondrocytes, therefore delaying the mineralization of the matrix.
The control of growth plate elongation is not a chondrocyte property, but a property of the growth plate module arising from the interaction with chondrocytes involved in the negative feedback loop of Ihh/PTHrP. Ihh also acts as a chondrocyte proliferation stimulator, through a PTHrP-independent pathway.
T-lymphocytes are also part of the regenerative process: They act by inhibiting the healing process through the action of cytokines IFN-γ and TNF-α.
MSCs have been reported to affect the immune response in a plethora of ways, through suppression or inhibition mechanisms. This response is coordinated by the cellular microenvironment and the MSCs-to-T-lymphocytes ratio, with a high ratio inhibiting the immune response, and a low ratio inducing it [40]."
According to XIAOLING LIAO, et al. [41]
"The bone marrow contains haematopoietic stem cells (HSCs) and mesenchymal stem cells (MSCs). The endosteum, located at the walls of the bone marrow cavity and the mesh of cancellous bones, contains the progenitor of osteoblasts (specialized cells capable of matrix secretion and mineralization) and osteoclasts (specialized in bone resorption). In the osseous tissue, there is abundant mineralized bone matrix and five kinds of cells: 1) Osteoprogenitors, including the progenitors of osteoblasts and osteoclasts; 2) Osteoblasts; 3) Osteocytes (a derivative of osteoblasts); 4) Osteoclasts; and 5) Bone-lining cells (inactive osteoblasts which can be re-activated by mechanical stimulus) [41-43]. Among these bone cells, osteocytes and bone-lining cells are the most abundant. The osteocytes are located in the spaces within the bone matrix, while bone lining cells cover all bone surfaces. The other three kinds of cells reside at the trabecular surface in cancellous bone or on the surface of the Haversian canals in the osseous tissues of compact bone [4,5]. Periosteum forms a layer outside of the osseous tissue. Both periosteum and endosteum play important roles in bone formation and growth and the bone remodeling process" [41].
Related the literature reported some facts can be observed: The high rate of regeneration in some invertebrates compared to adult of some advanced vertebrates. The difference showed in this last related age (since birth to later adult phases and ageing. The role played by innate and adaptative immunity and the finality that a regeneration present (competitive advantages in evolution pattern of life. The difference and similarity of process like wound repair, cancer and regeneration of a complete structure like lizard tail.
Complete reparative re-generation, as observed with limb re-generation in urodeles, is rarely encountered in mammals, being limited to re-generation after whole-thickness skin injury in certain species of mice. Other examples of physiological re-generation include replacement of red blood cells, epidermis, endometrium, and gut lining.
Invertebrates such as planarians and hydra, which can form whole animals from small segments, exhibit the greatest regenerative aptitude. Mammals, by contrast, fail to regenerate crucial structures, including limbs, spinal cord and cardiac muscle.
Certain vertebrates, including urodeles (e.g. salamanders) and teleost fish (zebrafish), retain the ability to regenerate these and other organs. Not only are lizards able to regrow cartilage and the spinal cord following tail loss, some species can also regenerate tissues after full-thickness skin wounds to the body, transections of the optic nerve and even lesions to parts of the brain.
Wound healing is an essential biological process involving the synchronized orchestration of numerous cellular and molecular events.
In lineage tracing experiments, new zebrafish cardiomyocytes have been found to be derived from de-differentiation of pre-existing cardiomyocytes to form an electrically coupled contractile syncytium.
Is interesting to observe the evolutive pattern involved in regenerative abilities across invertebrates and vertebrates and related the more complex body need (complexity, reproductive way, kind of immune systems: Innated, adaptative another).
Bergmann, using 14C dating and proved the occurrence of cardiomyocyte renewal in the human heart, with a yearly rate gradually decreasing with age from 1% at 20 years of age to 0.4% at 75 years of age. So the limited capacity of re-generation and proliferation of adult hearts still cannot compensate for the massive loss of cardiomyocytes in a single attack of MI.
Why adult miocardiocyte in humans lost their regenerative abilities?
There are some genetic reasons of this? When studying re-generation in reptiles and amphibians is "Why can these organisms regenerate, while mammals cannot?"
Comparisons between non-regenerative and regenerative organisms have identified 2 tissues/structures both unique to regenerative species and required for re-generation: 1) The apical cap; 2) The tail spinal cord; and 3) Distinct cellular mechanisms that can contribute to mammalian tissue re-generation after injury include:
1. Differentiation of recruited and/or resident stem and progenitor cell differentiation.
2. Replication of differentiated cells. This involves division of existing mature cells (hepatocytes) and can involve dedifferentiation of existing mature cells, proliferation and re-differentiation, as observed with re-generation of resected zebrafish hearts that results in almost complete structural and functional recovery, and in adult mouse heart following myocardial infarction-induced injury.
3. Transdifferentiation. This was initially observed for lens re-generation in the adult newt, where pigmented epithelial cells from the iris were found to transdifferentiate into lens cells. In mammals, re-generation via cellular transdifferentiation is observed in liver and pancreas.
Fibrosis occurs when there is a net increase in the rate of synthesis of the ECM. Excessive fibrosis can lead to increased left ventricular (LV) wall stiffness and decreased mechanoelectric coupling, adversely impairing cardiac contractile performance. In contrast, insufficient fibrosis in the heart post-MI can lead to LV wall thinning and rupture. Organisms that are not capable of tail re-generation, such as mice, do not possess tail spinal cords as adults, while both lizards and salamanders retain tail spinal cords into adult stages.
Reptiles and amphibians spontaneously regenerate cartilaginous skeletons in response to skeletal injury. The ability to regenerate whole appendages (limbs and tails) is a rarity among adult vertebrates. That destruction of spinal cords/ependyma at lizard tail amputation sites results in re-generation failure. It seems that interactions between the apical cap and tail spinal cord are as important to re-generation as the presence of these 2 tissues. The interactions are blocked by dermal tissue implants, and interposition of dermal tissue between tail spinal cords and apical caps inhibits re-generation. The most important tissue to re-generation is the tail spinal cord. Although re-generation is best known from salamanders and zebrafish, it is unexpectedly widespread among vertebrates. Not only are lizards able to regrow cartilage and the spinal cord following tail loss, some species can also regenerate tissues after full-thickness skin wounds to the body, transections of the optic nerve and even lesions to parts of the brain (This inverse relationship between skeletal development/maturity and re-generation fidelity, as well as the preference for producing cartilage, are also observed in tail re-generation). While many of the key mechanisms involved in wound healing (re-epithelialization, cell proliferation, angiogenesis, and extracellular matrix deposition and remodeling) are widely conserved, the fidelity of repair often varies.
In humans and most other mammals, non-lethal injuries typically result in the replacement of damaged tissues with a fibrous substitute known as a scar.
In contrast, other vertebrates including various species of bony fish (teleosts), salamanders and lizards are capable of wound healing without scar formation instead of replacing damaged tissue with a fibrous infill, these species undergo a tissue-specific program to restore tissue architecture and function. Although vertebrates lack the capacity for whole body re-generation, a broad range of organs can be partially replaced, including portions of the skin (epidermis and dermis), heart (ventricle), forebrain (telencephalon), spinal cord and even multi-tissue appendages, such as limbs and the tail. Fetal mammals can heal cutaneous wounds scar-free prior to the early- to mid-gestation period, while postnatal mice, rats, rhesus monkeys and human children can also spontaneously regenerate amputated digit tips another region of the central nervous system demonstrating variable responses to injury is the optic nerve.
In mammals and birds, damage to these axons can result in vision loss, as retinal ganglion cells degenerate and undergo cell death. Cellular de-generation and the inability to restore the visual pathway in these species appears to be the result of a complex inhibitory microenvironment, related to the formation of a glial scar (rich in proteoglycans and glial cells) and various axon impeding proteins such as Nogo-A.
As might be expected, species capable of restoring vision after injury to the optic nerve (zebrafish) are characterized by retinal ganglion cell survival, and the absence of axon inhibitory proteins such as Nogo and a glial scar.
Neotenic salamanders, which never fully develop and retain non-ossified, cartilaginous skeletons into adulthood, are able to regenerate fully formed limbs. With all the original cartilaginous skeletal elements of the originals these data reveal a surprising diversity across vertebrates in how the optic nerve responds to injury, with lizards uniquely interposed between full functional restoration and regenerative failure. How do lizards prevent (or at least limit) microbial invasion following tail or skin loss? Early evidence points towards the production of anti-microbial peptides (such as beta-defensins) as an important adaptation with obvious biomedical implications different mechanism of re-generation in the lizard than the blastema model described in the salamander and the zebrafish, which are anamniote vertebrates.
Molecular and cellular analyses in these model organisms are beginning to reveal conserved versus divergent mechanisms for tissue re-generation, which impacts the translation of these findings to human. An examination of the genetic regulation of re-generation in an amniote model will advance our understanding of the conserved processes of re-generation in vertebrates, which is relevant to develop therapies in humans. Amniote vertebrates, are evolutionarily more closely related to humans than other models of re-generation, e.g., salamander and zebrafish. The patterning and final structure of the lizard tail is quite distinct between embryonic development and the process of re-generation. Whereas the original tail skeleton and muscular groups are segmentally organized, reflecting embryonic patterning, the regenerated tail consists of a single unsegmented cartilaginous tube surrounded by unsegmented muscular bundles. Lizard tail regrowth involves the activation of conserved developmental and wound response pathways, which are potential targets for regenerative medical therapies. Through transcriptomic analysis of lizard tail re-generation, we have identified that genes in pathways involved in developmental processes, including myogenesis, Chondrogenesis, and neurogenesis, as well as adult processes, such as wound and immune responses, and are differentially expressed along the regenerating tail axis. Removal of the lizard apical cap, or replacement of the apical cap with mature skin grafts, inhibits tail re-generation.
The more skeletally matured lizards, on the other hand, regrow imperfect regenerated tails, and lizard cartilage tubes never segment and are easily distinguishable from original tail skeletons.
Transcriptomic analysis also revealed that genes involved in thyroid hormone generation were differentially expressed, suggesting a regulatory connection between re-generation of the lizard tail and Musculoskeletal transformations during amphibian metamorphosis. The lizarddio2 gene is the ortholog of deiodinase, iodothyronine, type I, which in mammals converts thyroxine prohormone (T4) to bioactive 3,3',5-triiodothyronine (T3). In Xenopus laevis, T3 is the key signal for the process of metamorphosis from tadpole to adult frog.
Many of the changes associated with metamorphosis are also observed in the remodeling of the tail stump and outgrowth of the lizard tail. No clear evidence of dedifferentiation has been identified in tail re-generation in the lizard, an amniote vertebrate. A temporal-spatial gradient of tissue patterning and differentiation along the regenerating tail axis has been described. It is possible that among the amniotes, the lizard retains genetic pathways associated with thyroid hormone regulation of metamorphosis in amphibian vertebrates. The re-generation of the amphibian limb involves a region of highly proliferative cells adjacent to the wound epithelium, the blastema, with tissues differentiating as they grow more distant from the blastema.
Re-generation requires a cellular source for tissue growth. The pattern of cell proliferation and tissue formation in the lizard identifies a uniquely amniote vertebrate combination of multiple developmental and repair mechanisms. Most also agree that the key limitation to cardiac muscle re-generation is likely to be the poor ability of adult mammalian cardiomyocytes to enter the cell cycle and undergo division.
Cardiomyocytes in the fetal mammalian heart are mononucleated and proliferative; but shortly after birth the vast majority of cardiomyocyte DNA replication occurs without cytokinesis or karyokinesis. Most cardiomyocytes are binucleated with diploid nuclei in the adult mouse heart, and mononucleated with polyploid nuclei in the adult human heart. After this postnatal switch, it is rare for cardiomyocytes to enter the cell cycle. Anversa and colleagues made use of human tissue samples collected from cancer patients who had received infusion of iododeoxyuridine, a thymidine analog used as radiosensitizer for therapy, and estimated that 22% of cardio myocytes in the human heart are renewed annually.
Salamanders are the champions of re-generation among vertebrates, able to renew removed or injured body parts like lens, retina, spinal cord, jaws, portions of intestine, brain tissue, and major appendages zebrafish; little or no collagen is retained by 1-2 months after resection injury. The clot is supplanted by cardiac muscle, restoring a contiguous wall of vascularized cardiac muscle.
Lizards are the only group of amniotes capable of tail re-generation as adults, and, unlike the anamniotic salamanders, adult lizard axial skeletons are fully ossified. Lizard tail re-generation follows waves of de-generation, proliferation, and differentiation.
Fibroblasts make up a high percentage of adult mammalian cardiac. Cells, and a much lower percentage of fetal mammalian or adult non-mammalian vertebrate hearts. These fibroblasts not only have the capacity to form scar tissue, but also appear to impact the proliferative capacity of cardiomyocytes. Study found that adult cardiac fibroblasts co-cultured with neonatal cardiomyocytes inhibited their proliferation, while embryonic cardiac fibroblasts had no such effect. Age-related changes in fibroblast characters might modify cardiac regenerative capacity. Relevant to cardiac cells, direct differentiation of non-cardiogenic mesoderm into beating cardiomyocytes and direct reprogramming of cardiac or dermal fibroblasts to cardiac muscle cells have been demonstrated. Mathematical modeling of the radiocarbon data suggested that human cardiomyocytes renew throughout life with a capacity that gradually decreases from ~1% annual turnover at the age of 25 to 0.45% at the age of 75. Amphibians survive massive mechanical injury to the ventricle, including removal of as much as one-quarter of the chamber. This resilience is a feat in itself, and is likely to reflect a lesser reliance than mammalian species on vigorous circulation.
In the newt heart, the eventual formation of connective scar tissue appears to be a dominant response after resection of the ventricular apex, and there is only minor replacement of cardiac muscle. Elevated indices of cardiomyocyte proliferation were detectable at the end of the first week after injury, and observable for weeks beyond this interestingly, these signs of heart failure reversed within several days, a recovery that correlated with massive cardiomyocyte proliferation detected throughout the ventricle. By 30 days after injury, the ventricle was filled with new muscle and displayed little or no scar tissue.
It is important to test whether regenerated cardiomyocytes incorporate functionally with existing cardiac muscle and do not generate arrhythmias. One possible reason for this may be intrinsic differences in cardiomyocytes. Lower vertebrate cardiomyocytes tend to be mononucleated, smaller in size, and containing fewer myofibrils, as compared to those of adult mammals. These characters are typical of cardiomyocytes in young mammals, and might facilitate cell cycle reentry after injury the capacity of myocardial re-generation is transiently present in the neonatal mouse heart, but is quickly lost by 7 days after birth.
Postnatal switches in cardiomyocyte proliferation and regenerative capacity coincide with changes of the expression of cell cycle regulator genes, and a recent study suggested the role of microRNAs (miRNAs) in this regulation. While cardiomyocyte characteristics would appear to have a primary role in regenerative capacity, another basis for the poor regenerative potential of the mammalian heart may be the activity of non-myocardial cardiac cells in response to injury. Injection of GFP-tagged cCFU-Fs into the infarcted heart demonstrated that those cells have the capacity to create cardiomyocytes, endothelial cells, and smooth muscle cells in vivo. The zebrafish endocardium quickly responds to injury and induces a signal(s) that is required for myocardial proliferation, while the endocardium of the adult mouse heart does not appear to mount an analogous response. Other candidate cardiac progenitor cells include "side population" cells that possess physiological properties to efflux fluorescent dye, or to form multicellular clusters, referred to as cardiospheres, in culture.
Colony-forming cells (cardiac resident colony forming units - fibroblasts, cCFU-Fs) in the adult mouse heart that have long-term growth potential in culture. Clonally derived cCFUFs were shown to give rise to multiple mesodermal lineages in vitro including cardiomyocytes, endothelial cells, smooth muscles, adipocytes, cartilage and bone. In adult mammals, Bersell, et al.: Neuregulin1 (NRG1) promotes proliferation of differentiated adult mouse cardiomyocytes in cell culture and when introduced in vivo. In the infarcted adult mouse heart, lineage tracing experiments indicated that the epicardium does not differentiate into cardiac muscle; instead, epicardial cells contribute to the canonical epicardial lineage (epicardium,fibroblasts, smooth muscle, perivascular cells). While data indicate that epicardial cells lack natural myogenic potential under most contexts, a recent study suggests that this restriction can be modulated [44].
Thymosin β4 (Tβ4) is a peptide that has been shown to enhance vascular potential to adult epicardial derived cells (EPDCs) and improve responses to MI. When Tβ4 was injected into mice prior to infarction, epicardial cells induced the expression of the embryonic epicardial gene Wt1 and cardiac progenitor markers. Within an hour of local injury, endocardial cells throughout the heart take on a rounded morphology and show detachment from underlying myofibers.
Concomitant with these morphological changes, endocardial cells induce the expression of developmental marker genes, raldh2 and heg, in an organ-wide manner by 3 hours postinjury. When neonatal mouse ventricles are injured by resection, cardiomyocyte mitoses and sarcomere disassembly are increased not only near the injury but also in areas distant from the injury. Various injury models suggest that the activation process in zebrafish does not require tissue removal or direct injury to the endocardium and epicardial tissue, and is not maintained by circulating systemic factors.
During mammalian liver re-generation, partial hepatectomy is known to affect tissue distant from trauma and activates compensatory hepatocyte proliferation in spare lobes, partly through inflammatory factors such as interleukin-6 and TNFα.
Members of the Fgf signaling pathway are upregulated after resection injury and may serve this purpose. Follow-up works found that expression of a receptor for Pdgf, pdgfrβ, is induced during heart re-generation, and that pharmacological inhibition of Pdgf receptors inhibits proliferation in epicardial cells and coronary vasculature formation during re-generation. Both Fgf and Pdgf signaling both appear to reactivate vascular development during myocardial re-generation. Cardiomyocyte dedifferentiation is typically characterized by reduction of sarcomere structures and expression of fetal gene markers, and appears to be a shared mechanism associated with cardiac muscle re-generation.
Braun, et al. recently investigated heart tissue samples from chronic dilated cardiomyopathy (DCM) patients, in an effort to discover factors that cause dedifferentiated phenotypes in human cardiomyocytes. By using proteomics and biochemical approaches, Oncostatin M (OSM) was found to be highly expressed in DCM hearts but not healthy hearts. OSM is a cytokine that has pleiotropic functions and transduces signals through a heterodimeric receptor composed of gp130, a co-receptor shared with many other cytokines, and OSM receptor (Oβ) or LIF receptor. OSM induced loss of sarcomeric structures and re-expression of embryonic markers in rat adult cardio-myocytes, through signals mediated by Oβ [45].
Invertebrates, ranging from protozoans to metazoans, possess cellular receptors, which bind to foreign elements and differentiate self from non-self. This ability is in multicellular animals associated with presence of phagocytes, bearing different names (amebocytes, hemocytes, coelomocytes) in various groups including animal sponges, worms, cnidarians, mollusks, crustaceans, chelicerates, insects, and echinoderms (sea stars and urchins). Basically, these cells have a macrophage-like appearance and function and the repair and/or fight functions associated with these cells are prominent even at the earliest evolutionary stage.
The primitive vertebrates (jawless fish) were the first to supplement innate responses with adaptive elements.
The high specificity, antibody maturation, immunological memory, and secondary responses of adaptive immunity were so successful that it allowed higher vertebrates to reduce the number of variants of the innate molecules originating from both invertebrates and lower vertebrates.
Organisms at all developmental stages have, in order to survive, applied available genes and functions of which some may have been lost or may have changed function through evolution. The molecular mechanisms involved in evolution of immune molecules, might apart from simple base substitutions be as diverse as gene duplication, deletions, alternative splicing, gene recombination, domain shuffling, retro-transposition, and gene conversion. Even prokaryotes protect themselves by use of restriction enzymes and clustered regularly interspaced palindromic repeats (CRISPRs), being able to degrade invading foreign pathogens. Since the appearance of the first eukaryotic cells, a series of additional defense mechanisms have evolved in order to secure cellular integrity, homeostasis, and survival of the host. Unicellular amebae developed the ability to Phagocytose foreign material as a part of their food uptake mechanisms [2] and this basic phagocyte function is conserved in higher invertebrates and vertebrates in which the immunological function is more evident. With the advent of cartilaginous and bony fish, the adaptive armament (MHC), immunoglobulins, T-Cell receptors, extensive cytokine ne2rks] appeared, and these New tools were further developed to a high level of sophistication. Through amphibians, reptiles, and birds to mammals. This Allowed a reduction of the copy number of many innate immune genes, but still the innate effector molecules have been taken into a complex ne2rk combining the obvious talents off a stacting ancient molecules with the highly developed specific recognition with memory seen in adaptive immunity. The main outlines of these aspects, which are presented below, high light how innate immune responses evolved from ancient precursors and still play a vital and basic role even in higher vertebrates where adaptive elements are so prominent." The regenerative capacity in tails or limbs progressively decreases in the frog as it transits from the larval stage to the post-metamorphic stage. This transition is closely linked to the maturation of the adaptive immunity. Regenerative programs demonstrated by lower vertebrates and young mammals have provided inspiring new insights.
In his 1935 treatise, Waddington considers the mechanistic connection between uncontrolled cancerous growth and controlled embryonic development, postulating the existence of "individuation fields" that regulate tissue growth both during embryonic development and in adult tissues. Modern interpretation of Waddington's theory, which remains untested and largely over-looked in the current literature, implicates re-generation mechanisms as possible cancer regulators and underscores the need to investigate links between re-generation and cancer. The term re-generation implies a well-coordinated restoration of cells, tissues, and organs that have been physically or functionally lost. This reparative process must accomplish the recognition and recapitulation of missing structures, while simultaneously achieving functional integration between recently formed and pre-existing tissues, in order to direct physiological and structural alterations. Re-generation involving cellular proliferation (epimorphosis) requires instructive signals with the capacity to efficiently regulate cell cycle, resulting in a finite number of cells that undergo division and complete repairs. The initial response would continue indefinitely, causing undesirable consequences for body homeostasis. Diverse regenerative phenomena appear to utilize similar mechanistic procedures, including: Cellular replacement (e.g., physiological cell turnover), local tissue repair (e.g., epithelial wound repair), and re-generation of large sections (e.g., appendages and head). The process is not infallible, as demonstrated by growing evidence associating re-generation with cancer-related cellular abnormalities. Seemingly contradictory, re-generation might in fact both contribute to the source of abnormal growth and also provide a means to prevent and correct growth abnormalities.
The second hypothesis conversely suggests that if induced cellular proliferation during re-generation is followed by morphogenetic processes, re-generation has the potential to prevent abnormal growth and more startling, to reverse malignancies and regain morphostasis-phenomena mostly observed in animals with immense regenerative potentials.
Biological responses associated with epimorphic re-generation lead to completely opposite outcomes- in one instance destruction, while in another rebuilding and reestablishing form and function. Even though urodeles and planarians both undergo epimorphic re-generation, the cellular mechanisms they use to repair missing parts and their sensitivities to carcinogens differ. While regenerative tissues in urodele amphibians display a low frequency of tumor development upon carcinogenic exposure, treatment of whole planarians with similar chemicals can lead to abnormal proliferation and tumor formation. The processes of lens and limb re-generation demonstrate an incredible degree of plasticity, where both differentiated pigment epithelial (PE) cells from the dorsal iris as well as limb mesenchyme reverse their differentiated state by re-entering the cell cycle. In both cases, this cellular dedifferentiation implies a loss of tissue-specific characteristics (bringing cells close to an undifferentiated state) followed by re-differentiation into cells of the same type or even a different lineage. This process can be repeated over and over without variation- a feature apparently missing in mammalian models where chronic local wound repair can lead to certain epithelial cancers. Adult tissues are able to reverse differentiated, post-mitotic cells to progenitor stages, thereby gaining migratory capabilities and massive proliferative properties that enable subsequent (re)differentiation, all without producing abnormal cell growth. Many of the results from these approaches were somewhat controversial, but they concluded that limb-regenerating tissue in urodele amphibians displays a very low rate of malignant tumor formation, such that these animals are commonly regarded as tumor resistant. Under certain conditions, malignant transformations (from the mucous glands of the skin) can be induced in newts after chemical treatment, confirmed by histological and pathological analyses showing epithelial tumors invading other tissues in patterns comparable to mammalian carcinomas.
In many cases malignancies were followed by spontaneous remission, as tumor cells reorganized into normal tissue (or sometimes abnormal but not cancerous structures). Just as significant are the similarities between cancer-originating cells and dedifferentiated limb cells. Planarians are free-living organisms of the phylum platyhelminthes well known for their capacity to regenerate entire animals from very small fragments. This invertebrate can increase its size by adding new cells or reduce it by removing cells, all depending on food availability.
Our current knowledge about cancer and abnormal outgrowth is largely derived from vertebrate works (particularly mammals), but similar growth aberrations have been reported in almost all major invertebrate phyla. Several works concluded neoblasts bear a functional resemblance to both adult mammalian stem cells and early-stage embryonic cells; particularly, neoblasts respond to carcinogenic agents by forming benign and malignant tumors, as well as teratomas in Dendrocoelum lacteum, a planarian species with limited regenerative capacities, where a differential response to chemically-induced carcinogenesis exists between regenerative (anterior) and non-regenerative (posterior) tissues. Anterior re-generation is capable of turning posterior infiltrating tumors into differentiated accessory organs such as the pharynx, which suggests the presence of regulatory long-range signals. Most planarian works seem to directly link chemical carcinogens to stem cell response, as neoblasts are the only adult proliferating cells in these organisms.
Comparisons of gene expression patterns between cancer and wound repair have revealed important differences in several pathways, from those associated with hypoxia-inducible factor and insulin-like growth factor-I, to genes regulating morphogenesis (e.g., CRYM, TCF21,CTGF, etc.) and glycolysis (e.g., PGK1 and HK1). Using RNA interference (RNAi) to downregulate an evolutionarily conserved tumor suppressor gene in S. mediterranea, Smed-PTEN, leads to abnormal neoblast proliferation and lethal phenotypes. An important regulator of the PI3K signaling pathway, PTEN is among the most commonly mutated genes in human cancer. In planarians, loss of PTEN function is characterized by tissue disorganization, disruption of basement membrane integrity (alteration of epithelial-mesenchymal interactions), and the presence of abnormal cells that invade distant tissues to form aggressive and eventually lethal ectopic outgrowths. Just as with carcinogenic treatments, re-generation could be inhibited in animals subjected to Smed-PTEN RNAi, indicating that under certain conditions PTEN is also required for re-generation.
A remarkable finding from these analyses was the striking regulatory conservation of the tumor suppressor function of planarian PTEN, which is similar to its mammalian counterpart but is absent in other invertebrates. The data suggest that molecular disruption of the PI3K signaling pathway in planarians results in abnormal stem cell proliferation, alteration of epithelial-mesenchymal interactions, and cellular infiltration into different tissues that altogether have been associated with vertebrate cancer development.
This also fits well with the previous hypothesis relating cancer to an incomplete regenerative process. Leukemia-initiating cells can be specifically abrogated in PTEN loss-of-function mice following rapamycin inhibition of the multicomplex protein TOR (a downstream component of the PI3K pathway). Components of the PI3K pathway are conserved in planarians, and rapamycin exposure following Smed-PTEN RNAi was similarly able to prevent phenotypic effects by specifically targeting abnormal proliferative neoblasts while maintaining the basal mitotic activity required for cell turnover.
In cases where virus-induced tumors (Lucke carcinomas) from anuran amphibians were implanted into actively regenerating newt limbs, tumor regression and subsequent differentiation into regenerating tissue was observed.
"Tumorigenesis can be considered as pathologically misappropriated tissue re-generation. Since most aggressive cancer cells mimic early-development stem cells, which properties of embryonic stem cells are retained in cancer cells? The first is related to cancer-progression-that cancer is a "wound that never heals"-that cancer metastasis embodies the concept of "seed and soil". The first concept refers to the fact that cancerogenesis and tissue re-generation are somewhat related processes and involve similar mechanisms, including i) Stem cell migration and recruitment and ii) The activity of chemotactic factors promoting cell motility. Cancer often originates in response to tissue/organ injury or chronic tissue inflammation, and evidence indicates the involvement of misappropriated homeostatic mechanisms that govern normal tissue repair processes and stem cell renewal. The "seed and soil" concept addresses the pro-migratory properties of cancer cells and their preferred pattern of metastasis to certain anatomical locations. The migratory potential of cancer cells mimics the mechanisms involved in migration of normal stem cells. Cancer cells respond to similar stimulating factors as do normal stem cells, follow gradients of similar chemo-attractants, and express a similar repertoire of adhesion molecules the unlimited proliferation potential of cancer-initiating cells mimics embryonic stem cells, with the major difference that malignant cells have defective differentiation potential.
"Although the regenerative response is complex, it typically involves the induction of new cell proliferation through formation of a blastema, followed by cell specification, differentiation, and patterning. Stem cells and undifferentiated progenitor cells play an important role in both tissue homeostasis and tissue re-generation" [46].
"Re-generation refers both to the regular and repeated renewal of a particular structure or tissue throughout the life of an organism, that is, the cellular renewal that occurs during normal aging (also called tissue homeostasis or physiological re-generation), as well as to restoration of injured tissue or lost body parts (also called reparative re-generation).
Importantly, the driver of physiological re-generation is replacement to maintain functional homeostasis, whereas reparative re-generation is triggered by injury signals [47]."
"Involvement of specific signaling pathways in lens re-generation, including the FGF, retinoic acid, TGF-beta, Wnt and hedgehog pathways [48]."
Stem cells are typically quiescent or passing slowly through the cell cycle in adult tissues, but they can be activated in response to cell loss and wounding. Regenerative capacity is regulated by a number of fundamental traits, including age, body size, life-stage, growth pattern, wound healing response and re-epithelialization, ECM dissolution (histolysis), re-innervation, and angiogenesis, as considered in detail for appendage repair.
A series of works, mostly performed in Drosophila as well as in Hydra, Xenopus, and mouse, has revealed an unexpected role of apoptotic caspases in the production of mitogenic signals that stimulate the proliferation of stem and progenitor cells to aid in tissue re-generation. Inflammation is commonly thought to promote tumorigenesis, and a complex relationship exists between inflammation and apoptosis. For example, hepatocellular carcinoma (HCC) frequently develops in response to chronic liver damage, and agents that induce cell death can promote HCC. The composition of the mammalian ECM also appears to be more inhibitory to cardiac re-generation than zebrafish ECM.
The chemical carcinogen DEN (diethylnitrosamine) induces hepatocyte death by DNA damage. DEN-induced hepatocyte death can activate the production of mitogens in adjacent myeloid cells, which in turn promotes compensatory proliferation of surviving hepatocytes cytokinesis, but not karyokinesis, in CMs is affected by matrix rigidity. Activation of both JNK and Stat3 has been implicated in several types of human cancer, and Stat3 function is critically required for the development of HCC. Because these pathways drive compensatory proliferation in Drosophila, and because caspase-3 and caspase-7 are required for liver re-generation in mouse, it is likely that caspases may regulate the production of some tumor-promoting cytokines. Inhibition of apoptosis contributes to the development and progression of cancer. Decellularized zebrafish ECM, prepared from normal or healing hearts, was shown to enhance functional recovery and myocardial re-generation when administered into the peri-infarct region of a mouse model of MI, as well as having pro-proliferative and chemotactic effects on human cardiac precursor cells in vitro. The enhanced regenerative response in the mouse appeared to involve erbB2 signaling as it was abrogated by inhibiting erbB2 activity.
In the nervous system, there are three types of ECM: A loose matrix present throughout the brain and spinal cord; matrix resulting from cell membrane-bound molecules; and a unique, lattice-like structure that wraps around specific neurons, called perineuronal nets (PNNs). PNNs are composed of highly negatively charged molecules, including hyaluronan, chondroitin sulfate proteoglycans, link proteins and tenascin R. Found in the nervous system of a variety of mammalian species, including humans, as well as in birds, such as zebra finch, PNNs limit plasticity in adulthood. The lateral elasticity of brain matrix is soft (Esoft ~1 kilopascal, kPa) compared to striated muscle, which is intermediate (Estiff ~10-17 kPa) and to osteoid, which is the heavily cross-linked collagen that initiates bone growth (Ehard ~20-50 kPa). Persistent inflammation leads to poor wound healing, with excessive fibroblast activity, ECM deposition and scarring.
After injury in the neonatal heart, for example, embryonic-derived resident cardiac macrophages expand, producing minimal inflammation; whereas, the adult heart contains embryonic-derived resident macrophages that are replaced by proinflammatory monocyte-derived macrophages after injury. Tissue resident macrophages in the mouse originate from one of three lineages, derived from yolk sac, fetal liver or bone marrow, which contributes to their diversity and allows them to play unique roles in organ development, homeostasis, and remodeling. Resolution of inflammation is essential, as prolongation of the inflammatory phase impairs re-generation and results in fibrosis, which impacts organ function. Even in moderately or highly regenerative species, such as anurans and urodeles, respectively, extending the inflammatory phase with pro-inflammatory beryllium sulfate treatment can cause limb re-generation to slow or fail completely. While an overactive immune response is detrimental to the regenerative process, species with reduced adaptive immune responses-that is, those leading to immunological memory that are activated by antigens and cytokines-have a greater capacity for wound healing and/or re-generation. Compared with the house mouse (Mus musculus), gerbils, African spiny mice (more closely related to gerbils than house mice), salamanders, nude mice (Foxn deficient), and xid (X-linked immunodeficiency) mice have an enhanced regenerative ability but are deficient in T- and/or B-lymphocytes.
In pro-regenerative species, such as urodeles and the African spiny mouse, it is clear that adaptive immunity and regenerative mechanisms are finely balanced to allow tissue repair.
Re-generation is dependent on an initial inflammatory phase for tissue debridement and protection from invading microbes. Tissue resident macrophages in the mouse originate from one of three lineages, derived from yolk sac, fetal liver or bone marrow, which contributes to their diversity and allows them to play unique roles in organ development, homeostasis, and remodeling. Resolution of inflammation is essential, as prolongation of the inflammatory phase impairs re-generation and results in fibrosis, which impacts organ function. While a population of monocytes and macrophages are recruited to the damaged tissue, each organ also has its own distinct resident macrophages, which are key to directing the injury response toward repair or fibrosis. While an overactive immune response is detrimental to the regenerative process, species with reduced adaptive immune responses that is, those leading to immunological memory that are activated by antigens and cytokines have a greater capacity for wound healing and/or re-generation. These works demonstrate that tregs are important modulators of the immune response across tissues with widely varying wound healing capabilities [49,50].
Further supporting the link between immune suppression and re-generation, anti-inflammatories have been used in a number of models to improve re-generation.
Mouse skin wound healing is improved and scarring reduced with celecoxib (a COX-2 selective non-steroidal anti-inflammatory agent) treatment.
Re-generation in tissues showing little or no self-renewal CNS. The CNS is composed of 2 major cell types: Neurons (electrically excitable cells responsible for transmission of information via electrical and chemical signals), and glial cells, which are divided into oligodendrocytes (responsible for myelination of axons), astrocytes (which interdigitate between neurons and blood vessels), ependymal cells (ciliated simple columnar cells that line the ventricles and central canal of the spinal cord) and microglia (resident macrophages responsible for immune defense in the CNS). In contrast to most branches of the animal kingdom, which show robust re-generation of their nervous system tissue, the CNS of vertebrates, including humans, has long been considered to be a "stable" or "perennial tissue", with little or no regenerative ability, the birth of neurons in the mammalian brain having been considered to be restricted to embryonic and early postnatal development. More recently, works using retrospective birth-dating based on the integration of atmospheric 14C into DNA also showed substantial turnover of adult human hippocampal neurons.
Analysis of 14C cell birth-dating indicates that ~35% of hippocampal neurons turn over at a rate of ~1.75% per year, and the rest are static. The functional role of these new hippocampal neurons in normal brains, let alone in disease, remains unclear. In mice, hippocampal neurogenesis mediates pattern separation in memory formation and cognition. Despite this, 14C cell birth-dating works provide evidence for the absence of postnatal neurogenesis in all major subdivisions of the human cerebral cortex (estimated turnover of 1 in 1000 neurons every 5 years, as well as in the cerebellum and olfactory bulb. This contrasts with robust adult neurogenesis in the olfactory bulbs of rodents and non-human primates. It appears that neither physiological nor reparative re-generation occurs in human cortical neurons, and there is also no evidence for tissue homeostasis in the olfactory bulb or cerebellum.
A number of molecular pathways are involved in regulating adult mammalian neurogenesis, including Wnt, Notch, epidermal growth factor (EGF), sonic hedgehog (Shh), bone morphogenetic protein (BMP) and a series of neurotrophic factors. Works of optic nerve re-generation in reptiles indicates that unlike fish, these animals do not recover vision even with axonal regrowth to the tectum, unless they undergo visual conditioning. In this situation, factors, such as increased brain complexity appear to underlie differences in functional recovery despite similar adequate reparative re-generation. Fundamental differences are observed between regenerating species, such as salamanders and larval frogs versus non-regenerating mammals, in the response of radial glial cells, which have proliferative and neurogenic capacity after spinal cord injury. Only in the former does the response of radial glia, through epithelial to mesenchymal transition, allow reconstitution of the spinal cord neuroepithelial tube that is critical for complete regenerative repair.
In mammals, glial cells respond differently, infiltrating the wound and depositing ECM. This leads to glial scar formation, which initially stabilizes the tissue and prevents further damage by necrotic cells, but ultimately impedes axonal extension through the injury, rather than allowing reconstruction of the spinal cord tube or damaged brain. Hence, fundamental differences exist in the properties and responses of glial cells in different animal phyla, which will likely continue to challenge efforts to affect repair clinically in response to nervous system injury.
To maintain high pressure in a closed circulatory system for adequate organ perfusion, the mammalian heart has evolved as a robust contractile organ. This requires adequate embryonic and postnatal development, the latter involving a marked increase in its mass over a relatively brief period (almost fourfold in 25 days between postnatal days 10 (P10) and P25) to adapt rapidly to increases in circulatory demand. The myocardium is highly vascularised with capillary beds originating from the left and right coronaries, supporting the high demand for coronary blood flow and oxygenation, with a capillary-to-myocyte ratio of 1:1. Of the three major cardiac cell types: CMs, endothelial cells, and fibroblasts, CMs account for 65-85% of the myocardial mass, but only 20-33% of the total cell population of the mammalian heart. Endothelial cells and fibroblasts are actively renewed throughout life, with predicted annual turnovers of ~17% and ~4% in the adult human heart, respectively. Lineage tracing works, 14C birth-dating works, non-radioactive nucleotide incorporation works, and stereological CM counting works indicate that CM turnover is detectable in the young but declines rapidly with age (~0.76% per year in mouse; 1% per year at age 25 years falling to 0.45% per year at age 75 years in humans) [51,52].
In the mouse, Cardiomyocytes (CM) generation has been reported to be restricted to a small fraction (< 0.2%) of mononucleated CMs. This study could not rule out contributions from binuclear CMs.
It is now generally agreed that postnatal CM generation is due to proliferation of existing CMs, rather than to maturation of stem/precursor cells. The number of CMs increases postnatally in the rat and mouse. Continued postnatal CM generation is consistent with a greater increase in heart weight than in body weight in the period immediately before adolescence, which is due to a surge in circulating thyroid hormone (T3) levels as the hypothalamic/pituitary/thyroid axis matures and also due, in part, to hypertrophic CM growth during this time. An increase in CM population number was also observed in humans during the first 20 years of life, from 1 billion at birth to 4 billion in adults. Design-based stereology to quantitate CM numbers showed that CM generation in human hearts is robust in early childhood but then declines, so that of the ~40% of CMs generated throughout life, only 3% are "born" after age 10 years. Surprisingly, CM numbers remained constant throughout life (3.1 billion CMs at birth and in adults), implying that the robust CM generation observed during infancy is not due to continued postnatal growth of the heart but, rather, to replacement of lost CMs, although cell death was not evaluated in this study. Repair of the embryonic mouse heart following ablation of 50-60% of cardiac progenitor cells or immature CMs is complete with full re-generation [53].
Histologically, the myocardium regenerates with resolution of scar/clot and increased DNA proliferation, and function of the intact heart is restored. New CM proliferation involves an increase in the proliferation rate of immature CMs, above the already brisk proliferation rate observed at this time in uninjured hearts. This level of replacement is almost on par with that observed in zebrafish, where 70-80% of lost CMs can be replenished. Complete cardiac re-generation, evident by complete histological repair and clot resolution, is observed in neonatal mice following resection of the LV apex, MI due to occlusion of the left anterior descending coronary artery, cryoinjury or genetic CM ablation.
Re-generation following apical resection or MI is limited to a brief developmental window, being robust when effected in P1 hearts but not P7 hearts; a time when CMs have exited the cell cycle and are quiescent. Consistent with CM re-entry into the cell cycle during preadolescence, myocardial ischemic (MI) injury at this age leads to a partial regenerative response. This is evident by decreased scar size, increased BrdU labeling and improved ejection fraction compared to mice experiencing MI injury at a later age (P21). In the neonatal mouse infarct model, re-generation has been shown to involve not only CM replication but also robust angiogenesis and revascularisation. CM renewal is driven by division of pre-existing CMs. Repair of the heart following injury has been suggested to involve CM dedifferentiation, division and redifferentiation. Dedifferentiation is characterized by disassembly of sarcomere structure, extrusion of mitochondria, electrical uncoupling, and expression of precursor cell markers and of regulators of cell cycle progression.
Re-differentiation involves restoration of cell morphology, sarcomere organization and contractile function. Adult CMs subjected to ischemia undergo dedifferentiation, proliferation and re-differentiation as evidenced in an in vitro co-culture model (adult CMs co-cultured with neonatal rat ventricular myocytes), as well as in post-infarct hearts. Ischemia induces gap junction uncoupling in the periinfarct zone as a result of hypoxia-mediated dephosphorylation of the gap junction protein, connexin 43-the major mediator of intercellular communication, including propagation of calcium transients.
Liver mass is restored within 7-10 days in rodents and after 6-8 weeks in humans. Unlike re-generation of the resected liver in zebrafish, which undergoes epimorphic re-generation, the normal architecture of resected adult mammalian liver lobes is not regenerated in this way. Rather, repair after hepatectomy involving up to 30% of the liver mass, in the adult rodent, is achieved by hypertrophy of the remaining hepatocytes in all residual lobes, resulting in increased metabolic activity without hepatocyte division. As in the heart, lineage tracing works of hepatocyte proliferation in both rats and mice indicate that stem cells are not involved in restoring adult liver mass. Cardiac re-generation with full functional recovery has been reported in several case works of infants and children afflicted with diphtheria or after a perinatal infarct. Scarless repair of the myocardium has also been observed after corrective surgery for a congenital cardiac anomaly. The repair process appears to be coordinated by liver sinusoidal endothelial cells (LSECs), which activate vascular endothelial growth factor receptor 2 (VEGFR-2) and tyrosine kinase with immunoglobulin-like and EGF-like domains 2 (TIE-2) signaling, resulting in the secretion of angiocrine factors (Wnt2 and hepatocyte growth factor), and cytokines (CXCR4 and CXCR7) that trigger hepatocyte proliferation and liver repair. Hepatic stellate cells (liver pericytes) are also activated to secrete hepatocyte growth factor and hedgehog, which stimulate hepatocyte proliferation.
Hepatic macrophages upregulate Wnt signaling in response to phagocytosis of dead cells. In contrast to the adult liver, 20-30% hepatectomy in neonatal (day 0.5) mice results in numerous rounds of clonal cell division and full reconstitution of lobe architecture. This is similar to the re-generation observed in the neonatal heart Liver repair following 70% hepatectomy is associated with dynamic changes in the expression of specific miRNAs, such as miR-21, miR-221, and miR-26a [54,55].
These miR changes correlate with changes in the expression of target genes that play important roles in liver repair, such as those encoding growth factors or cell cycle regulators.
Recent works also indicate a role for long non-coding RNAs (lncRNAs) in liver repair, examples being LALR1 (lncRNA associated with liver re-generation 1) and MALAT1 (metastasis-associated lung adenocarcinoma transcript 1). These lncRNAs promote cell cycle progression and accelerate hepatocyte proliferation by activating Wnt/β-catenin signaling. Based on current evidence, the plasticity of differentiated cells contributes to tissue repair in the liver under both homeostatic and injury conditions, with cholangiocytes acting as facultative liver stem cells to effect repair when hepatocyte regeneration is impaired.
Physiologically, β-cell proliferation in humans and rodents occurs at a low level (1-3% of human cells and 10-30% of murine cells in cell cycle) in neonates and the early stages of life, after which there is an age-dependent decline in β-cell proliferation (to ~0.1-0.2%). Adult human β-cells fail to respond to the same growth factors and nutrients. Human β-cells do not proliferate during pregnancy or in response to insulin resistance. In adult human β-cells, most of the key G1/S molecules, such as cyclins and cyclin-dependent kinasese (CDKs), are not found in the nucleus, but in the cytoplasm. Replicative quiescence in β-cells might be due in part to the inability of cyclins and CDKs to access the nuclear compartment and this may be a common feature of other non-proliferating adult cells, such as skeletal muscle, CD8+ memory T cells, keratinocytes, and prostate gland cells. miRNAs regulate β-cell proliferation. One example is miR-7a, which inhibits adult mouse and human β-cell proliferation via inhibition of the mTOR pathway. Another example is histone modifications through trimethylation of H3K27 or H3K4 that are regulated by Polycomb or Trithorax group protein complexes, respectively.
In juvenile rodent and human β-cells, this restricts access to promoters of genes encoding cell cycle inhibitors, thereby permitting β-cell proliferation, but these histone modifications are reduced in adult human β-cells, thereby restricting proliferation. Unlike the liver, the pancreas does not have bona fide stem/progenitor cells [56].
Lessons from fetal models and highly regenerative species demonstrate that control of inflammatory responses, rapid ECM remodeling and the deposition of collagen type III are necessary for complete scarless re-generation, suggesting a line of investigation for the development of therapies to improve skin re-generation."
Even in the same individual, different tissues or organs exhibit distinct regenerative capacity; livers regenerate more readily than hearts in mammals. Stem or progenitor cells. The activation of stem/progenitor cells is the most popular way to generate new cells; it is reasonable to assume that the abundance of stem cells, to some extent, reflects the regenerative capacity.
As we mentioned above, planarians, Hydra, and Xenopus laevis possess a large number of pluripotent, multipotent, or unipotent stem cells. Not surprisingly, they all have the high regenerative capacity. In adult mammals, a small number of tissue-specific stem cells are preferentially preserved in certain high-turnover tissues. Animals or tissues with more stem cells generally possess higher regenerative capacity. Dedifferentiation potential primitive vertebrates such as salamanders and zebrafish still regenerate substantial parts of their body, even without the presence of numerous stem cells. That is because they can produce new cells easily via dedifferentiation. Compared with zebrafish cardiomyocytes, adult human cardiomyocytes retain a limited ability to enter the cell cycle: A very low level (0.0006% to 1%) of constant cardiomyocyte turnover rate occurs throughout life.
Mammals fail to regenerate bones after the amputation of their bones, although internal bone defects can be healed below a critical size. In mammals, de novo osteoblasts deriving from mesenchymal stem cells contribute to the bone-healing process, without the occurrence of osteoblast dedifferentiation. Although mammalian cells are hard to take natural dedifferentiation after injury, dedifferentiation can be induced in vitro.
Mouse myotubes are induced to dedifferentiate and proliferate after treatment with extracts from regenerating limbs of newts or after ectopic expression of the transcription factor, msh homeobox 1 Msx1. These indicate that mammalian cells (like myotubes) remain the potential to dedifferentiate, although the potential needs to be stimulated. Terminally differentiated newt myotubes can dedifferentiate after injury because tumor suppressor retinoblastoma (Rb) proteins are phosphorylated, thereby allowing cells to re-enter the cell cycle. Mammalian myotubes do not phosphorylate Rb proteins after injury and therefore fail to re-enter the cell cycle. Rb and another Rb family member p130 can block cell-cycle genes and maintain the postmitotic state of mammalian adult cardiomyocytes; knockdown of Rb and p130 leads to the cell-cycle re-entry of adult cardiomyocytes.
As a cell-cycle inhibitor, the p53 tumor suppressor also hinders dedifferentiation. During salamander limb re-generation, an early down-regulation of p53 is a prerequisite for mesenchymal cell dedifferentiation and blastema formation. Because cell-cycle inhibitors block dedifferentiation in mammalian cells, targeted modification of these inhibitors is likely to promote dedifferentiation and re-generation. The forced expression of transcription factors or treatment with small molecules changes the epigenetic regulators such as DNA methylation and histone modification, resulting in the complete dedifferentiation of somatic cells into pluripotent stem cells. The loss of natural transdifferentiation in mammals appears to impede complete lens re-generation. Nonetheless, mammalian cells retain the transdifferentiation potential, which has to be incited by exogenous stimuli. The latent transdifferentiation in mammals is extensively confirmed by recent reprogramming strategies.
Somatic cells, such as fibroblasts, can be induced into another lineage (e.g., neurons, cardiomyocytes, and hepatocytes) by several reprogramming approaches, including lineage factor-based reprogramming, induced pluripotent stem cell (iPSC) factor-based reprogramming, and small molecule-based reprogramming.
The eukaryotic genome is packaged into chromatin consisting of DNA, histones, and nonhistone proteins. The chromatin structure has profound effects on gene expression, because it regulates the accessibility of transcription factors and transcriptional machinery to their target DNA. Chromatin can be remodeled as loose chromatin (euchromatin) or dense chromatin (heterochromatin) via epigenetic regulators, including DNA methylation, post-translational modifications of histones (e.g., acetylation and methylation), and ATP-dependent chromatin remodeling.
Dynamic changes in chromatin states result in the increased or decreased expression of genes. DNA methylation is the most studied epigenetic regulator, including cytosine methylation (5 mC) and cytosine hydroxymethylation (5 hmC). In general, high levels of DNA methylation repress gene expression and low levels of DNA methylation promote gene expression. The low DNA methylation pattern in the genome appears to closely associate with the regenerative capacity. Low DNA methylation is observed in the MRL/MpJ mouse model, which exhibits an enhanced regenerative response in a variety of organs, including livers, ears, and hair follicles. Some genes related to embryonic morphogenesis, such as EPH receptor A2 (Epha2), paired box gene 2 (Pax2), and GATA zinc finger domain containing 2A (Gatad2a), are hypomethylated and highly overexpressed in the adult MRL/MpJ mouse.
At the cellular level, DNA methylation status correlates with dedifferentiation potential. Xenopus tadpoles can regenerate a full limb after amputation, whereas Xenopus froglets (young frogs) can form only a simple cartilaginous spike structure after amputation. Although they both can form a blastema upon amputation, the froglet blastema fails to regenerate. The deficient expression of the re-generation associated sonic hedgehog (Shh) gene in Xenopus froglets leads to loss of re-generation.
The enhancer region of the Shh gene is highly methylated in the Xenopus froglet blastema and thereby silenced, but it is hypomethylated in the Xenopus tadpole blastema as well as in the salamander blastema. Planarian Schmid tea mediterranea histone deacetylase 1 (Smed-HDAC-1) that is specifically expressed in neoblasts also maintains the stem property of neoblasts.
Depletion of the SET/MLL or the Smed-HDAC-1 leads to the loss of planarian re-generation.
Injured neurons in the peripheral nervous system (PNS) can successfully regenerate axons, whereas neurons within the central nervous system (CNS) typically fail to regenerate axons after injury.
Adult dorsal root ganglion (DRG) neurons after the peripheral axotomy show increased active H4 acetylation surrounding the axon re-generation genes, leading to their expression.
DRG neurons after a central lesion fail to increase H4 acetylation, accompanied by no expression of those genes; when H4 acetylation is increased by administration of an HDAC inhibitor in the mouse model of spinal cord injury, axon re-generation is significantly improved. Peripheral nerve injury can trigger nuclear export of HDAC5 whereby HDAC5 levels are reduced in the nucleus. Reduced nuclear HDAC5 level, in turn, increases histone acetylation at the re-generation promoting gene loci and activates their transcriptional expression.
All the works demonstrate that different epigenetic responses to injury (such as histone acetylation) may lead to a discrepancy in regenerative capacity between PNS and CNS. Histone modifications are also associated with aging-related loss of regenerative capacity.
In livers of older mice, the CCAAT/enhancer-binding protein alpha (C/EBPα)-HDAC1 complex accumulates in the region of the E2F-dependent promoters of liver proliferation-associated genes, thereby suppressing these genes and reducing the regenerative capacity of older livers.
The immune system is implicated in tissue homeostasis and wound repair. Meanwhile, the inflammatory interactions of immune cells and fibroblasts often bring about scarring or fibrosis. Comparative analyses of animal re-generation display an inverse relationship between the evolution of the immune system and the regenerative capacity.
On the contrary, salamanders can regenerate limbs completely, whereas frogs fail to regenerate limbs, indicating that regenerative capacity declines as the immune system advances.
Zebrafish have a higher CNS re-generation than mammals have, which is associated with a much weaker and shorter inflammatory response to CNS injury in zebrafish than in mammals.
Observing evolution of invertebrates and vertebrates is possible to verify that regenerative of tissue and organ in more expressed in more ancients organism and that in more advanced animals this abilities is showed only in the first phases of their life.
Other consideration: Evidences say us the more advanced animals with adaptative immunity seem have reduced regenerative properties.
The same it is interesting to observe that the introduction in evolution of some structure like bone skeleton in a more rigid body produced a modify in regenerative need for the organism itself.
Genetic evolution play a great role ad conserved genes can be verified also in animals who lost this regenerative abilities (see regenerative abilities in some animals in first stage of their life).
Some difference can be also compared between regeneration of a total organ or tissue and a repair wound or a neoplastic formation: In regeneration a complete genetic program seem able to start and stop the process without over production or uncontrolled increase.
An interesting last consideration: Is seem that advanced organism with more complex structure then invertebrates and with adaptative specific immunity and with complex rigid organization need less regenerative abilities: In example it is not useful a over regeneration I a body with or inside a rigid structure because an overproduction could result I ineffiecient anatomophisiology.
Finally is possible to consider that the introduction of rigid or semirigid bone structure was linked with a reduction of phenotipic expression of regeneration genes?
The regenerative genes are conserved gene among animal evolution, but what event is related to the silentiating of this? Some phenotypic expression in non-regenerating adults animals is due to a suppression of genotypic conserved genes (inibithing factors).
Is seem that a genetic program produce in the same time inibithing signal and related loss of regenerating abilities. And what kind of negative signal was involved? But if this mechanism is useful in an organism very complex and with more rigid skeleton is the same useful in all conditions? This negative influx is relevant in some therapeutic setting?
In example in transplant activity (bone marrow, regeneration of retina or after IMA)?
Immune responses are, critically involved in regulating the wound healing process and must be temporally and spatially controlled for epimorphic re-generation to occur.
Related reference citated is possible to verify that in bone tissue many inhibiting factor are present so it is possible to say that this tissue can be considered not a good environment for stem cells logarithmic expansion?
What role played in evolution bone structure introduction? Bone physiology implies various phases of re-adsorbtion and production of new tissue in a balanced way. So it is easy to think in activating mechanism but also a stopping one genetically mediated.
We have see advanced animals with bone skeleton show reduced regeneration abilities and this is and real interesting fact. A rigid structure need less regenerative abilities because more little space of action to replace tissue I the same volume.
In exoskeleton in fact there are limited possibilities to increase body complexive volume and this confirm this theory.
I regeneration a genetic message imply a start and a stop message to reproduce the tissue - structure to be replaced following a codified pathway.
In this process also the ending message are crucial. But what happen I stem cell transplant if this inibithor messages complex is reduced or deliminated? In example using out of bone environment?
It is possible to say that it is important not only the way of regeneration but also why? What microenvironmental factors were involved to silentiating of some genetic pathway in evolution of animal life.
This research work is produced without any therapeutic or diagnostic intent and only to produce new research hypothesis.