In this study, we evaluated the stability of three common housekeeping genes, GAPDH, beta-Actin, and 18S-rRNA, in human neuroblastoma SHSY-5Y cells exposed to varying levels of steroid hormones like progesterone (P4) and testosterone. Housekeeping genes serve as reliable internal controls, assumed to maintain consistent expression across diverse tissues and experimental conditions. Since steroid hormones act as ligand-activated transcription factors, directly impacting gene transcription, the identification of suitable housekeeping genes in steroid hormone-influenced tissues is imperative.
Our analysis, employing the Norm finder, Genorm, and Best Keeper algorithms within the web-based tool Ref Finder, indicates that GAPDH emerges as the most stable housekeeping gene among the three candidates. In contrast, 18S-rRNA is deemed the least suitable, showing significant up regulation with low doses of progesterone treatment and down regulation in cells treated with testosterone and high doses of progesterone. It is worth noting that the ribosomal fraction might not accurately represent the overall cellular mRNA population.
Acknowledging the study's limitations, including the focus on SHSY-5Y cells with unique metabolic characteristics, we recommend cautious extrapolation to different cell types, tissues, and hormone concentrations. Determining an appropriate housekeeping gene is vital before embarking on new experiments. This study serves as a foundation for future research in qPCR analysis, shedding light on how steroid hormones influence commonly used housekeeping genes. It prompts the need for further exploration of hormone effects in diverse cellular contexts, contributing to enhanced research on life-imparing diseases.
SHSY-5Y, Progesterone, Testosterone, Steroid hormones, PCR, Housekeeping genes
Selecting appropriate housekeeping genes to normalize data is essential when applying RT-qPCR since their expression varies depending on the tissue and treatment and the choice of housekeeping gene will ultimately influence statistical interpretation of data. Steroid hormones are known to be ligand activated transcription factors in cell nuclei and therefore have a direct influence on the transcription of various genes [1]. Hence, evaluating RT-qPCR data of such steroid-hormone treated tissue is challenging. The significant variation observed in housekeeping genes under diverse experimental conditions has the potential to result in a misinterpretation of the expression pattern of a target gene [2]. So far, no suitable housekeeping genes for human neuroblastoma cell-line SHSY-5Y treated with progesterone or testosterone have been reported.
In addition to its major role in female reproductivity, progesterone is known to be a neurosteroid of the central (CNS) [3-5] and peripheral nervous system (PNS) [6,7], inducing neuroplasticity, neuroregeneration and neuroprotection [8,9]. Two studies recently showed that progesterones neuroprotective properties even extend to the enteric nervous system (ENS) [10,11]. To this date, very few studies addressed possible housekeeping genes regarding the influence of progesterone on housekeeping genes. The neuroprotective potential of the gonade steroid testosterone has only recently been discovered. With publications found on pubmed doubling in the last 20 years from 4000 publications in 2004 to over 8000 publications per year in 2022. So far, only one study addresses the influence of testosterone on the expression of housekeeping genes [12]. SHSY-5Y is a human neuroblastoma cell line and a sub line of the SK-N-SH cell line, which was first established in culture in 1970 from a biopsied bone marrow metastasis of a 4-year-old female [13]. SHSY-5Y cell-line exhibits catecholaminergic and neuronal properties and is widely used in in vitro studies about neurodegenerative and neurodevelopmental disorders, such as Parkinson’s Disease, Alzheimer’s Disease and even autism spectrum disorder [14-16]. This particular cell line is commonly selected due to its human origin, catecholaminergic neuronal traits, and its user-friendly maintenance demands [17]. Nevertheless, up to this date few publications about suitable housekeeping genes when working with SHSY-5Y are available.
As the levels of testosterone and progesterone vary depending on the cycle and biological sex [18,19], vulnerability of the selected HK gene to these hormones in each tissue should always be considered when comparing groups of different sexes. With this study we aim to provide the reader with a suitable HK gene for such implications.
Figure 1 gives an overview over the experimental setup. Exponentially growing SHSY-5Y neuroblastoma cells were seeded in T25 tissue culture flask at a density of 1 × 104 cells/cm 2 at day 1. The cells were maintained in a humidified atmosphere at 37 °C and 5% CO 2 in proliferation medium (Table 1). 24h after plating, the proliferation medium was replaced with differentiation medium containing retinoic acid to promote differentiation and induction of a neuronal phenotype. After 48h the differentiation medium was replaced by differentiation medium containing the external factors corresponding to the assigned group (progesterone (1 nM, 10 nM, 100 nM), testosterone (1 nM, 10 nM, 100 nM). As described in [11] progesterone was diluted in EtOH until the desired concentration was reached. Note that the final concentration of EtOH in the medium was never higher than 1%, which is described to have no influence on cell integrity [20]. After an incubation period of 24 hours adherent cells were collected using cell-scrapers followed by centrifugation for 5 min at 4 °C and 1000 rpm. Each experiment was performed independently three times and all three samples were used for further processing and analysis.
 Figure 1: Timeline of the experiments.
View Figure 1
Figure 1: Timeline of the experiments.
View Figure 1
Table 1: Compounds used for media and treatments. View Table 1
After discharging the supernatant, the pellet was used for RNA extraction using the Nucleo Spin RNA-Kit (740955.50, Macherey-Nagel, Düren, Germany). The RNA was eluted twice using the same 40 µl of nuclease-free water to increase the yield. The eluted RNA was measured by the Nano Drop™ One/One C ND-ONE-W, Thermo Scientific™ to ensure optimal quality and quantity for downstream applications. cDNA synthesis was performed using the Go ScriptTM Reverse Transcription Mix, Oligo(dT) (#A2790, Promega, Madison, WI, USA) with a total input of 1 µg of RNA. Both kits were used according to the manufacturer’s protocol.
In the following RT-qPCR the expression levels for the candidate housekeeping genes were measured in duplicates and in three independent experiments. 5 µl GoTaq qPCR Master Mix (#A6001, Promega), 2.9 µl nuclease-free H 2 O, 0.5 µl Primer upstream, 0.5 µl Primer downstream and 2 µl diluted cDNA were combined to a reaction volume of 10 µl per well. The cDNA was diluted at 1:15 in nuclease-free H2O. Using the CFX96 Real Time PCR Detection System (Bio Rad, Hercules, CA, USA) samples were heated to 95 °C for 2 min, followed by 40 amplification cycles, 15s at 95 °C and 60s at 60 °C. For each primer pair, dissociation curves to confirm PCR amplification specificity and exclusion of unspecific PCR products and primer dimers were performed (Table 2).
Table 2: Used primers for quantitative real-time PCR. View Table 2
In a first attempt the achieved Ct values were analyzed as raw data. For further investigations a standard curve to calculate primer efficiency was run. The Ct values were then normalized to the differentiated but untreated cells also integrating the primer efficiency. Additionally qPCR data (Ct values) were analyzed using three widely established algorithms: Bestkeeper [21], NormFinder [22] and Genorm [23] on RefFinder web-based comprehensive tool [24,25]). The Ct value was determined by the count of cycles required for the fluorescence to attain a particular threshold detection level. This value exhibits an inverse relationship with the quantity of template nucleic acid existing in the reaction [26].
The employed programs all use a different algorithm to determine the stability of the candidate housekeeping gene. Best Keeper is Excel-based and uses pair-wise correlation analysis of candidate genes. After determining the best-suited housekeeping gene out of up to ten candidates the software combines them into an index. In this software all data processing is based on crossing points.
Norm Finder is also Excel-based algorithm for identifying the optimal normalization gene among a set of candidate genes. Ct-values cannot be used directly and must be transformed into linear scale expression quantities. This can be done by a standard curve or the delta-Ct method. The software calculates inter- and intra group variability among the tested samples. It provides a stability value for each gene which is a direct measure for the expression variation as estimated by the software.
Genorm is part of Biogazelle’s qbase+ software for quantitative PCR data analysis and older implementations remain available online, for python or R. to program executes on the base of non[1]normalized expression levels and calculates pair wise variation for every control gene with all the other control genes and defines the average pair wise variation of each particular gene as the internal control[1]gene stability measure M. The lower the M values, the more stable the expression of the candidate gene.
The RT-qPCR analysis of the expression patterns of three candidate housekeeping genes GAPDH, beta-Act in and 18S-rRNA confirmed differential expressions following treatment with different doses of steroid hormones.
Upon examination of the mean cycle threshold (Ct) values (Figure 2), it is evident that 18s and Actin exhibit closely similar expression levels, with mean Ct values of 18.23 and 18.05, respectively. In contrast, GAPDH presents a slightly higher Ct value, detected at 19.36. This suggests that GAPDH is expressed at a relatively lower level compared to the other two housekeeping genes under the tested conditions. However, a more comprehensive assessment considering the standard deviation (SD) highlights a different aspect of the results. Both GAPDH and Actin demonstrate significantly lower SD values, measuring 0.34 and 0.30, respectively. This indicates that these two genes exhibit higher stability in their expression across the experimental conditions. On the other hand, 18s-RNA exhibits a notably higher SD value of 0.62, suggesting that its expression is less stable and more variable under the same conditions.
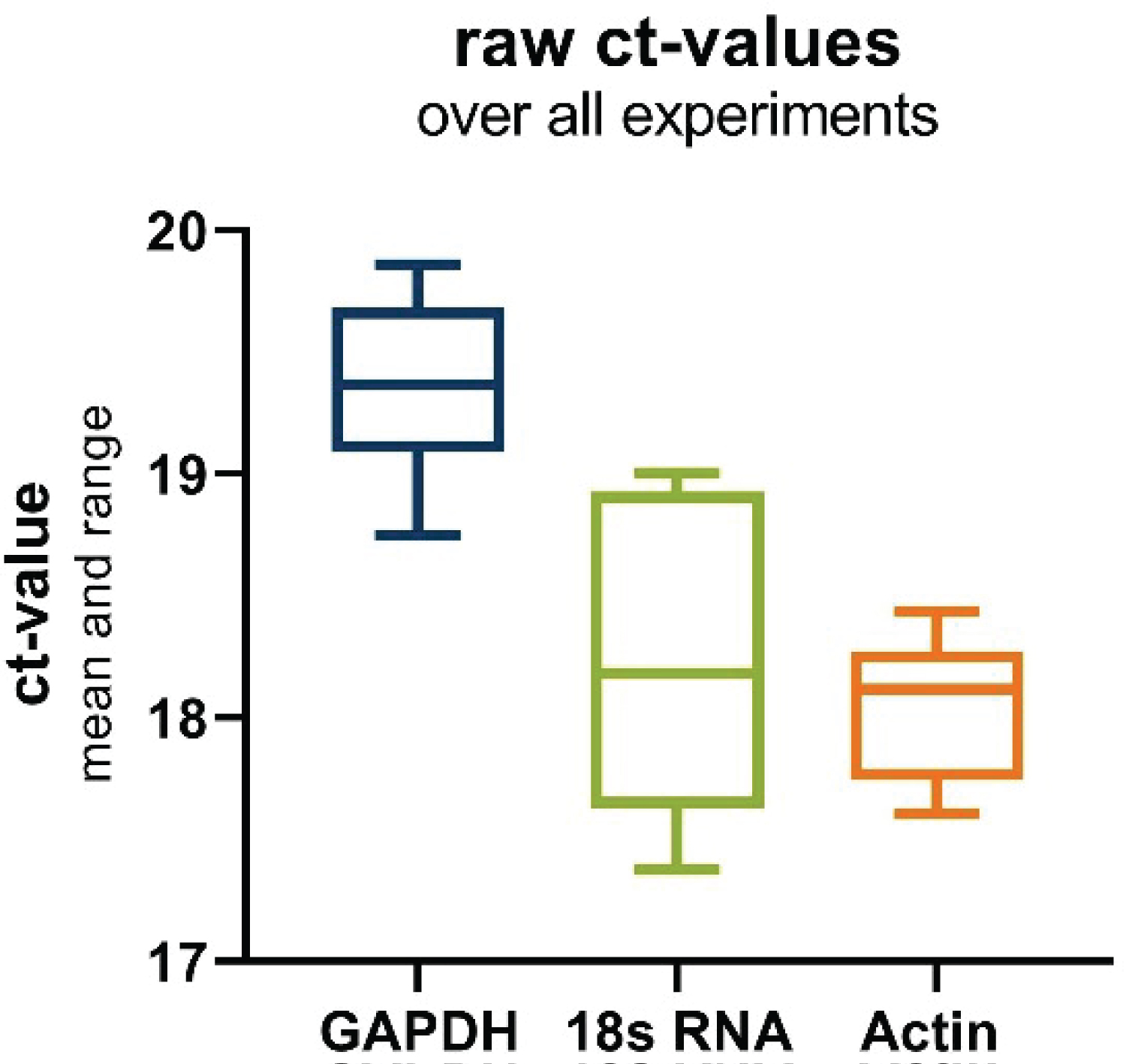 Figure 2: Boxplot of raw ct-values of the investigated housekeeping genes over all experiments. Depicting the range and mean.
View Figure 2
Figure 2: Boxplot of raw ct-values of the investigated housekeeping genes over all experiments. Depicting the range and mean.
View Figure 2
In the next set of observations (Figure 3), it becomes evident that steroid hormone treatment exhibits a tendency to influence the expression of housekeeping (HK) genes, with distinct patterns emerging among the tested genes. For GAPDH and Actin, a general trend towards decreased gene expression is observed as progesterone concentrations rise. At a progesterone concentration of 1 nM, the raw cycle threshold (Ct) values for GAPDH and Actin are approximately 19.1 ± 0.2 and 17.7 ± 0.5, respectively. In contrast, at a higher progesterone concentration of 100 nM, the Ct values are approximately 19.7 ± 0.3 (GAPDH) and 18.4 ± 0.6 (Actin). These findings suggest a potential down regulation of GAPDH and Actin expression in response to progesterone, albeit without statistical significance.
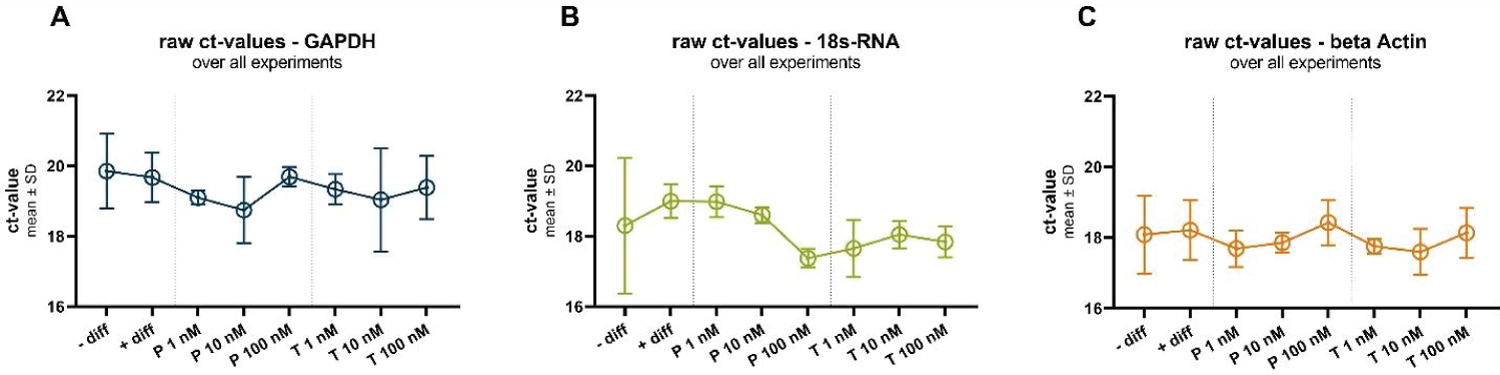 Figure 3: Raw ct-values of all three investigated housekeeping genes GAPDH (A), 18s-RNA (B) and beta Actin (C), under all investigated conditions: In undifferentiated (-diff) and differentiated (+diff) cells and under different progesterone (P) and testosterone (T) treatments.
View Figure 3
Figure 3: Raw ct-values of all three investigated housekeeping genes GAPDH (A), 18s-RNA (B) and beta Actin (C), under all investigated conditions: In undifferentiated (-diff) and differentiated (+diff) cells and under different progesterone (P) and testosterone (T) treatments.
View Figure 3
Conversely, 18s RNA displays an opposing trend, as increasing progesterone concentrations appear to up regulate its expression. This is evident in the Ct values, where at 1 nM progesterone, the Ct value for 18s RNA is approximately 19.0 ± 0.4, and at 100 nM progesterone, the Ct value is approximately 17.4 ± 0.3. Again, it is important to emphasize that these trends do not achieve statistical significance, but they offer valuable insights into the potential regulatory effects of progesterone on 18s RNA expression.
Similar trends are observed in response to testosterone treatment, further highlighting the consistency of these patterns across different hormonal interventions.
Figure 4 displays the standard curves for the examined housekeeping genes. Utilizing linear regression analysis, we derived the equations that represent these curves, which, in turn, were employed to ascertain the primer efficiencies. The resulting equations are as follows: Y = -3.283X + 15.15 (for 18s RNA); Y = -3.368X + 16.24 (for GAPDH); Y = -3.878 * X + 17.83 (for Actin). Consequently, these equations facilitated the computation of the percentage efficiencies of the primers, yielding primer efficiencies of 99% for GAPDH, 81% for Actin, and 102% for 18s RNA. These calculated primer efficiencies played a pivotal role in generating the data that have been normalized to the differentiated cells, as depicted in the subsequent figure (Figure 5).
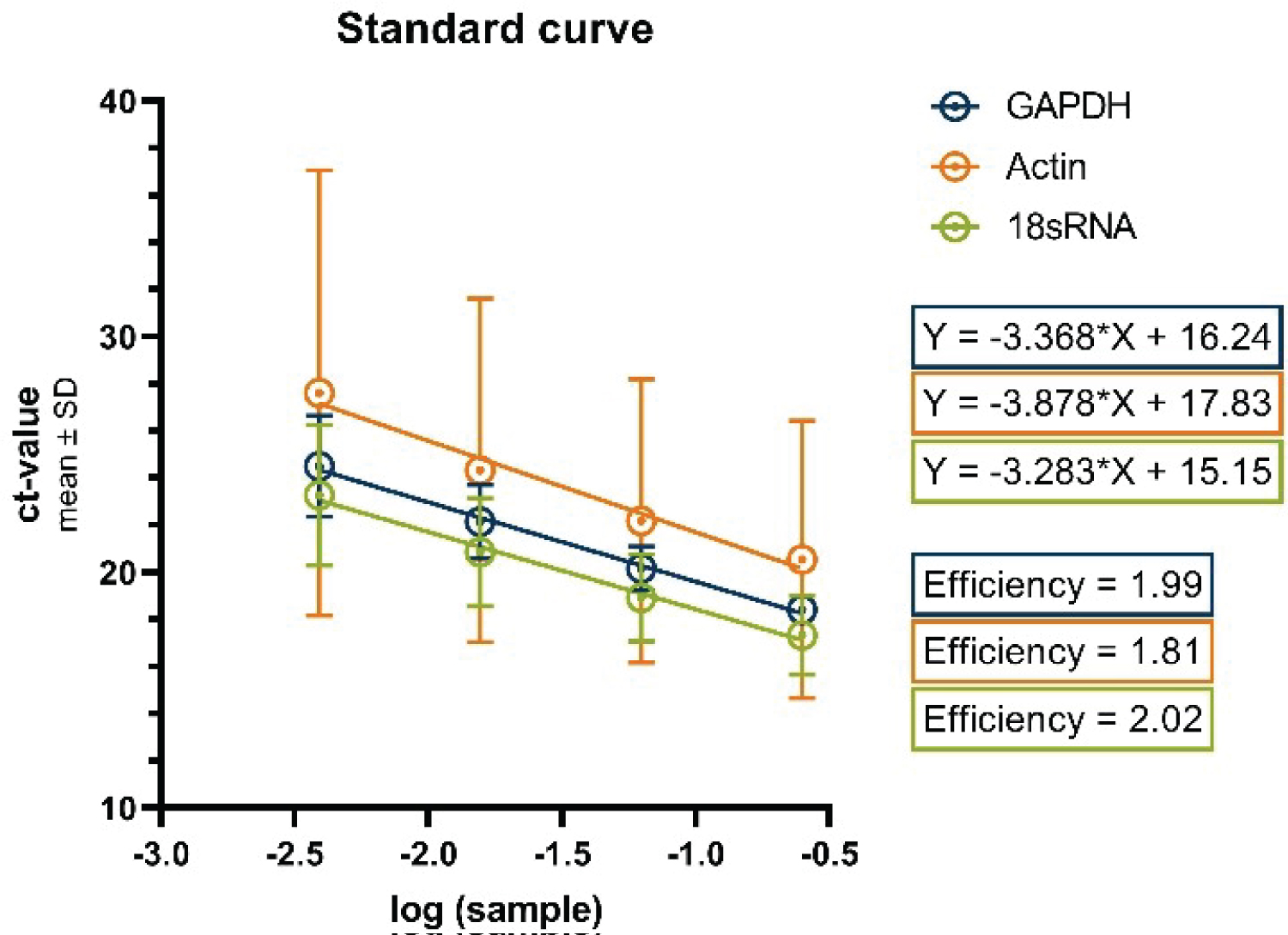 Figure 4: Standard curves for the housekeeping genes GAPDH, Actin and 18s RNA.
View Figure 4
Figure 4: Standard curves for the housekeeping genes GAPDH, Actin and 18s RNA.
View Figure 4
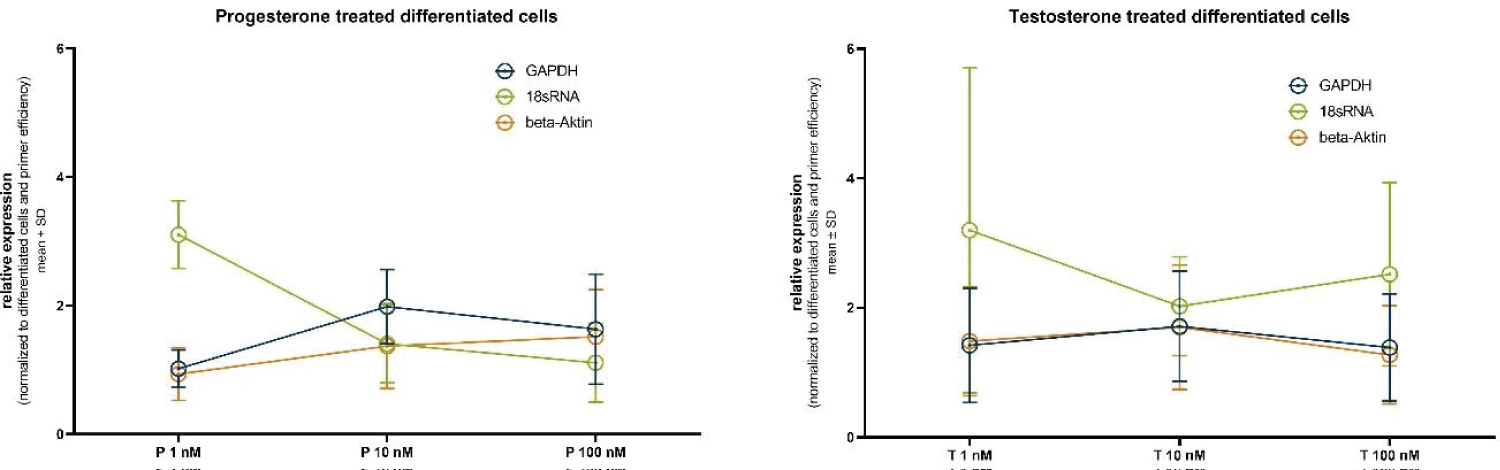 Figure 5: Relative housekeeper gene expression after normalization to the differentiated untreated cells and treatment (A) progesterone or (B) testosterone.
View Figure 5
Figure 5: Relative housekeeper gene expression after normalization to the differentiated untreated cells and treatment (A) progesterone or (B) testosterone.
View Figure 5
The incorporation of these standardized curves and calculated primer efficiencies serves to enhance the precision and reliability of our gene expression analysis. Furthermore, it enables the generation of normalized data, thus affording valuable insights into the expression levels of the housekeeping genes under investigation within the context of differentiated and steroid hormone treated cells.
Figure 5 presents the normalized relative expression of the selected housekeeping genes following treatment with different concentrations of progesterone (1 nM, 10 nM, and 100 nM).
For GAPDH, the relative expression after 1 nM progesterone treatment is 1.02 ± 0.29, at 10 nM it is 1.98 ± 0.58, and at 100 nM it is 1.63 ± 0.86. These results indicate a slight increase in GAPDH expression with higher progesterone concentrations, although statistical significance is not achieved.
In the case of 18s RNA, the relative expression at 1 nM progesterone is 3.10 ± 0.53, at 10 nM it is 1.41 ± 0.61, and at 100 nM it is 1.11 ± 0.61. Notably, 18S-rRNA exhibits a substantial decrease in expression with increasing progesterone concentration, again without reaching statistical significance.
Similarly, Actin displays variations in relative expression levels. At 1 nM progesterone, it is 0.93 ± 0.41, at 10 nM it is 1.37 ± 0.66, and at 100 nM it is 1.52 ± 0.74. While there is a discernible trend of increased Actin expression with higher progesterone concentrations, statistical significance remains elusive.
In a parallel set of experiments mirroring the progesterone treatment, we evaluated the normalized relative expression of housekeeping genes following treatment with different concentrations of testosterone (1 nM, 10 nM, and 100 nM). For GAPDH, at 1 nM testosterone, the relative expression is 1.42 ± 0.88, at 10 nM it is 1.71 ± 0.85, and at 100 nM it is 1.39 ± 0.83. These results indicate a moderate fluctuation in GAPDH expression with testosterone concentration, though statistical significance remains absent. In the case of 18s RNA, at 1 nM testosterone, the relative expression is 3.20 ± 2.51, at 10 nM it is 2.02 ± 0.76, and at 100 nM it is 2.52 ± 1.42. Similar to the progesterone treatment, 18s RNA exhibits substantial variations in expression levels, albeit without reaching statistical significance.
Likewise, Actin demonstrates changes in relative expression levels. At 1 nM testosterone, it is 1.48 ± 0.84, at 10 nM it is 1.70 ± 0.96, and at 100 nM it is 1.27 ± 0.76. Here again, a trend of altered Actin expression with varying testosterone concentrations is observed, without achieving statistical significance.
The Norm finder algorithm rates GAPDH as the most stable gene in this context with a stability value of 0.211 followed by beta-Actin (stab. value .435) and 18S-rRNA as the least stable gene with a stability value of 0.76. In the ranking according to Best Keeper algorithm rates GAPDH as the candidate gene with the highest stability followed by beta-Actin and 18S-rRNA. Genorm suggests a combination of GAPDH and beta-Actin as the most suitable normalization gene and 18S-rRNA as the least stable gene. While the employed algorithms agree on the gene stability ranking, they differ in the magnitude of the difference (Figure 6).
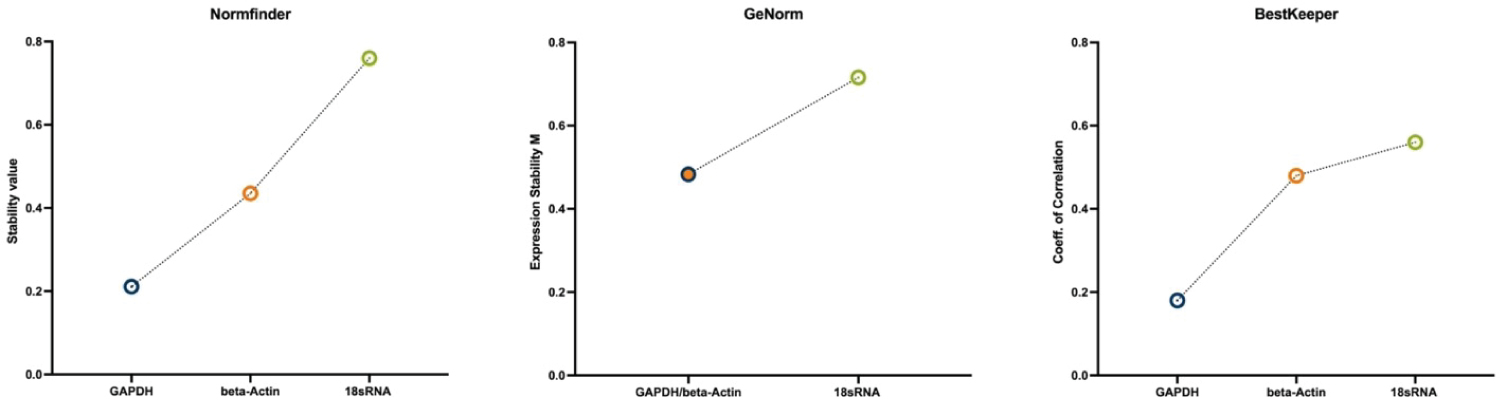 Figure 6: Stability ranking of the three used algorithms, showing most stable gene on the left to least stable gene on the right. All three algorithms rank GAPDH (alone or in combination) as the most stable expressed gene.
View Figure 6
Figure 6: Stability ranking of the three used algorithms, showing most stable gene on the left to least stable gene on the right. All three algorithms rank GAPDH (alone or in combination) as the most stable expressed gene.
View Figure 6
In the present work the stability of the candidate housekeeping genes GAPDH, beta-Actin and 18S-rRNA in human neuroblastoma SHSY-5Y cell line treated with different levels of steroid hormones progesterone and testosterone was evaluated using the algorithms Norm finder, Best keeper and Genorm as they are implemented in the web-based tool Ref Finder. When performing rt-qPCR, it is essential to control for experimental variations in the amount of RNA used in each experiment and the efficiency of the reverse transcription reaction. To achieve this the expression of control genes that are supposed to have a stable expression between different tissues and are meant to be unaffected by experimental treatments are used. Steroid hormones are known to act as ligand-activated transcription factors in cell nuclei and therefore have a direct influence on the transcription of various genes [27]. Therefore, it is crucial to identify suitable housekeeping genes in steroid hormone-affected tissues.
Three of the most commonly employed housekeeping genes are Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), beta-Actin and ribosomal 18S-RNA. GAPDH is an enzyme of glycolysis and catalyzes the reversible oxidative phosphorylation of glyceraldehyde-3-phosphate [28]. Beta-actin is a major cytoskeletal protein with a universal expression across various cell types. It is among others involved in cell division, cell migration and phagocytosis [29]. 18S-rRNA is one of the basic components of all eukaryotic cells as it is a component of the ribosomal small subunit (40 S subunit), which is indispensable for protein biosynthesis [30].
Our data suggest that GAPDH is the most suitable housekeeping gene among the three selected candidates as proposed by algorithms like Norm finder, Genorm and Best Keeper. In contrast to this, 18S-rRNA has shown to be the least appropriate housekeeping gene in steroid hormone treated tissue, as it proved to be significantly upregulated in cells treated with low doses of progesterone and down regulated in cells treated with testosterone and high doses of progesterone. In addition, the ribosomal fraction might not represent the overall cellular mRNA population [31].
As so far no research on the influence of progesterone and testosterone on the expression of GAPDH, beta-Actin and 18S-rRNA in human neuroblastoma SHY-5Y cells has been published, it remains difficult to evaluate our data in the context of the present literature. But our data are in line with previous findings of Crythorn, et al. 2009, who found that the presence of progesterone has an influence on 18S-rRNA regulation in the uterus of mice. Similar to us Crythorn, et al. observed an increase in 18S-rRNA expression in the progesterone treated samples [32].
In primary cultures of human thyroid cells treated with progesterone and estradiol Norm finder software identified beta-actin as the most stable housekeeping gene, followed by Tata-Box binding protein (TBP) and GAPDH [33]. These finding are similar to our results, as the Norm finder software gave similar stability values to GAPDH and beta-Actin. Nevertheless, it has to be kept in mind that due to different metabolic and general properties, the comparison of different cell types is not always valid.
In line with our results, testosterone and low doses of progesterone (< 100 nM) were shown not to have an effect on GAPDH enzyme activity in rat brain solubilized plasmalemma-micorosomal or cytosolic fractions [34].
In the context of real-time PCR (rt-PCR) analysis, the associated formulas (e.g.: 2-∆∆ct) assume a 100% primer efficiency (where 2 equals 100%). This can introduce a potential source of error, as demonstrated by the findings in this study, which indicate variability in primer efficiency. Rychlik, et al. [35] observed that the stability of both the primer and target DNA duplex is influenced by the GC content within the primer sequence. Furthermore, Pan, et al. [36] highlighted the preference of specific primer motifs by DNA polymerases, a preference that can vary between different polymerases and primer sequences. A study conducted by Bustin, et al. [37] emphasized the impact of a poor primer design and the failure to optimize reaction conditions, which can result in reduced technical precision and the flawed detection of amplification. According to Bustin, et al. commonly used primers in published assays may lack the intended specificity, exhibit dimer formation, possess a narrow temperature range for hybridization, or compete with template secondary structures. Consequently, it is strongly recommended to perform primer efficiency analyses with the chosen (housekeeping) primers and the DNA polymerase intended for use in future experiments.
Care should be taken when applying our results to different experimental set-ups implying other cell, tissue types or hormone doses as the cell's responses to hormonal treatment may vary. Based on the presented data we highly recommend to establish a suitable housekeeping gene prior starting an experiment. Therefore this paper aims to provide a comprehensive overview of how steroid hormones, progesterone and testosterone, exhibit their influence on different commonly used housekeeper genes and can function as foundation for future research with accurate results in qPCR analysis.
Lastly, we would also like to discuss the limitations. Although three repetitions were carried out for each test condition, there is a notably standard deviation in some cases, which means that the data must be interpreted with caution. The algorithms itself also contain pitfalls, as, for example, a 100% primer efficiency is assumed, which, as shown, does not always correspond to real scenarios.
The study focused primarily on human neuroblastoma cells SHY-5Y, which are known for their altered metabolism. While interventions influenced by progesterone and testosterone are promising, external validity should be critically evaluated. Further investigation into the effects of various hormones commonly found in the organism on housekeeper genes is warranted. Advances in technology and a more thorough exploration of the role of hormones in cellular processes will provide a broader understanding and contribute to more accessible research on life-impairing diseases.
All in all, we encourage fellow researchers to publish their experiences with different HK genes and make their knowledge accessible to the community, as our study underscores the need to choose appropriate housekeeping genes for different experimental setups and cell lines.
This research received no external funding.
The authors wish to thank Felicitas Opdenhövel, Claudia Grzelak and Anke Lodwig for excellent technical assistance. FORUM & Heinrich-Alma-Vogelsang Stiftung.
The authors declare no conflict of interest.