A new immunohistochemistry (IHC) method using fluorescence-emitting phosphor-integrated dot (PID) nanoparticles as a fluorescent dye was developed to accurately detect proteins that are expressed by cancer cells. This technique localizes proteins and quantitates their levels of expression in cancer lesions, which facilitates objective analysis of protein levels. We used the PID nanoparticle assay to measure HER2 expression in formalin-fixed, paraffin-embedded (FFPE) samples from patients with primary breast cancer and evaluated the suitability of this new technique for clinical use.
We performed IHC with PID nanoparticles (IHC-PID) using FFPE samples from 41 patients with primary breast cancer to quantitatively assess HER2 expression compared with conventional HER2 assays: IHC and in situ hybridization (ISH).
The PIDs/cell was significantly higher in HER2-positive compared with HER2-negative patients, independent of the type of assay. There was a significant correlation between the in situ hybridization (ISH) ratio and PIDs/cell (Spearman rank correlation r = 0.4833, P = 0.0014). Receiver operating characteristic curve analyses revealed that the measurements of PID nanoparticles predicted HER2 expression. When we determined HER2 status using only ISH, the cut-off value was 3.1 PIDs/cell (sensitivity and specificity, 100% and 55%, respectively), area under the curve = 0.843.
Quantitation of HER2 protein expression using PID nanoparticles shows promise for preclinical and clinical application to breast cancer.
Breast cancer, HER2, Protein expression, PID, Nanoparticles
HER2: Human epithelial growth factor receptor 2; IHC: Immunohistochemistry; ISH: in situ hybridization; DISH: dual-probe in situ hybridization; PID: phosphor-integrated dot; IHC-PID: IHC using PIDS; FFPE: formalin-fixed paraffin-embedded; ASCO/CAP: American Society of Clinical Oncology/College of American Pathologists; PBS: phosphate-buffered saline; PFA: paraformaldehyde-phosphate buffer solution; ROI: region of interest; ROC: receiver operating characteristic; AUC: area under the curve
Human epithelial growth factor receptor 2 (HER2, also designated ERBB2) is a member of the epidermal growth factor receptor family and the target of anti-HER2 antibodies such as trastuzumab. HER2 is amplified, overexpressed, or both in 10%-25% of human breast cancers and is associated with aggressive metastatic disease and poor prognosis [1]. HER2 expression is typically assessed using immunohistochemistry (IHC), and the HER2 copy number is determined using an in situ hybridization (ISH) assay. IHC is easier to perform, although results can vary if the sample conditions or staining processes differ, such that classifying HER2 status is subjective. ISH is often performed when IHC analysis classifies HER2 status as equivocal, because the ASCO/CAP HER2 testing guideline states that IHC 2+ (equivocal) cases must be reexamined for the HER2 status with a different tissue block, if available, with the same HER2 testing method or by an alternative HER2 testing approach with the same tissue block [2]. Furthermore, ISH achieves better diagnostic accuracy and added confidence, although it is more time-consuming, labor-intensive, and expensive compared with IHC.
A new IHC method using fluorescence-emitting phosphor-integrated dot (PID) nanoparticles as a fluorescent dye, called High Sensitive Tissue Testing, was developed to accurately detect proteins expressed by cancer cells [3-7]. PIDs are uniform particles in size, 149 nm in average. According to a fluorescence spectral analysis, 580 nm light-excited spectral patterns of PID particles peaked at approximately 620 nm in wavelength. A single PID has approximately 2,460 streptavidin molecules linked via polyethylene glycol (PEG) chains on its surface, and thus, they highly react to the secondary antibody [6]. PIDs coupled to target proteins (antigens) are detected using a fluorescence microscope and then analyzed using specialized software [7]. PIDs exhibit high luminance and a high dynamic range of fluorescence. Therefore, IHC using PIDs (IHC-PID) is more sensitive compared with methods that employ conventional fluorescent dyes [6]. Moreover, this technique localizes proteins and quantitates their levels of expression in tumor lesions.
Here we performed IHC-PID to quantitate HER2 levels in formalin-fixed, paraffin-embedded (FFPE) samples from patients with primary breast cancer. We evaluated whether this new technique can be translated to the clinic.
We used IHC-PID to analyze 41 patients with primary breast cancer, analyzed in our previous study [8], who underwent surgery at the University of Tokyo Hospital from 2009 to 2011 (Table 1). Patients with ductal carcinoma in situ, a tumor < 1 cm, and history of chemotherapy were excluded. The median age of patients was 60.0 years (range, 28-85 years). Tumors ranged from 10 mm to 43 mm. All samples included in the study were previously assessed for HER2 status according to the guidelines of the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) [2]. A tumor was considered HER2-positive using the criteria as follows: 1. The IHC score was 3+ positive (circumferential membrane staining that is complete, intense, and detected in > 10% of tumor cells) or 2+ positive (circumferential membrane staining that is incomplete, weak/moderate, or both in > 10% of tumor cells, or complete), and intense circumferential membrane staining detected in ≤ 10% of tumor cells). 2. The HER2/CEP17 ratio was ≥ 2.0, or average HER2 copy number was ≥ 6.0 signals/cell determined using fluorescence in situ hybridization or differentiation induction subtraction hybridization, counting ≥ 20 cells within the area. A tumor was considered HER2-negative using the criteria as follows: 1. The IHC scored 0 (no membrane staining), 1+ (faint or barely perceptible/incomplete membrane staining), or 2+. 2. The HER2/CEP17 ratio was < 2.0, and the average HER2 copy number was < 4.0 signals/cell determined using ISH. None of the samples analyzed here was an ISH-equivocal case (HER2/CEP17 ratio < 2.0; average HER2 copy number ≥ 4.0, and < 6.0 signals/cell). Among 10 HER2 3+ samples, four were judged HER2 2+ at the time of diagnosis, according to published guidelines [9]. Fifteen patients were diagnosed here as HER2-positive. Among 15 HER2 positive cases, according to either ASCO/CAP 2013 or 2007, 7 cases received anti-HER2 therapy and 8 did not (Table 2).
Table 1: Clinicopathological characteristics of patients included in study (N = 41). View Table 1
Table 2: HER2 status and PID nanoparticle counts.
Among HER2 positive cases diagnosed according to ASCO/CAP 2007, seven cases (with 〇) received anti-HER2 therapy and six cases (with ×) did not. The sign of (*) of cases 29 and 37 next to 〇 means that these cases had recurrence of breast cancer within three years after surgery.
Italics applied to cases 28 and 35 means that PIDs/cell were very low considering their IHC score (3+ according to ASCO/CAP 2013) and ISH (> 2.0); The cells applied with grey means that PIDs/cell varied among the spots.
View Table 2
HER2 IHC staining was carried out on FFPE tissue using Ventana BenchMark XT (Roche Diagnostics) with the 4D5 anti-HER2 antibody. Staining intensity and membrane immunoreactivity patterns were evaluated using the Dako scoring system according to ASCO/CAP guidelines [2,9].
HER2-DISH was performed on FFPE tissue using the HER2 DNA DISH kit (Roche Diagnostics) according to the manufacturer's instructions. Specimens were evaluated by light microscopy. HER2 signals appeared as black dots or clusters, and CEP17 signals appeared as red dots. Twenty non-overlapping cancer nuclei were scored for HER2 and CEP17 signals and HER2 gene amplification was classified according to ASCO/CAP guidelines [2,9].
IHC-PID examinations were performed using the same blocks as those for IHC and ISH.
Specimens were fixed in 10% formalin and embedded in paraffin, cut into 3-µm-thick sections, and placed on glue-coated glass slides (Matsunami Glass, Osaka, Japan). Sections were deparaffinized in xylene and hydrated with distilled water. Antigen retrieval was performed in citrate buffer (pH 6.0) at 95 ℃ for 40 min, and the samples were then cooled at room temperature for 40 min. After washing three times with phosphate-buffered saline (PBS) for 5 min, sections were blocked by incubation with B-001 (Konica Minolta, Tokyo, Japan) at room temperature for 15 min. The sections were reacted with a primary antibody against HER2 (Pathway anti-HER2/neu (4B5) Rabbit Monoclonal Primary Antibody; Roche Diagnostics, Mannheim, Germany) at 4 ℃ overnight. Sections were incubated with the secondary antibody (2 µg/mL) (SA-001; Konica Minolta, Tokyo, Japan) for 30 min and then with PID-conjugated streptavidin (0.03 a.u; Konica Minolta) for 2 h at 25 ℃. To visualize nuclei, sections were washed with PBS, fixed with 4% paraformaldehyde-phosphate buffer solution (PFA; Wako Pure Chemical Industries Ltd., Osaka, Japan) at room temperature for 10 min, and then incubated with Mayer's hematoxylin solution (Sakura Finetek, Tokyo, Japan) at room temperature for 1 min. The sections were washed with distilled water, dehydrated, cleared, and mounted.
A pathologist selected five spots randomly within invasive components of a tumor in each section. For each spot, a 174.4-µm × 130.8-µm field (microscopic field of view at ×400 magnification) was defined as a region of interest (ROI) (Figure 1). In each ROI, fields containing only nonmalignant cells, such as stromal cells or fibrous tissue, were manually trimmed if necessary, in order to exclude noises in the area of nonmalignant cells and to decrease the possibility of error counting of PIDs. In each spot, number of PID particles and number of cells are counted, and average PIDs per cell is calculated (PIDs/cell = [total PID counts in the spot]/[total cell counts in the spot]). Fluorescence emitted by the PIDs was detected using an epifluorescent microscope (BX53; Olympus, Tokyo, Japan) equipped with a camera (DP73; Olympus, Tokyo, Japan), and analyzed using imaging software (cellSens standard version 1.12; Olympus, Tokyo, Japan). Filters used for laser-excited fluorescence and exposure times are described in a previous report [3]. Image analysis software (PidAnalyzer 2.14.1.1; Konica Minolta) and image editing software (Image J version 1.48; Konica Minolta) were used for data analysis [4].
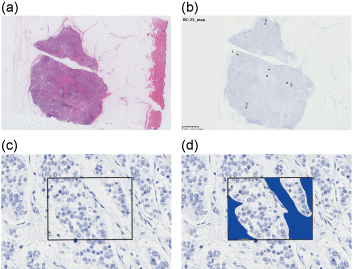 Figure 1: ROIs selected for image analysis a) Hematoxylin-eosin-stained section of sample 22; b,c) Five spots within a tumor were selected, and a 174.4-µm × 130.8-µm field was defined as an ROI for each spot; d) In each ROI, fields without tumor cells, such as stromal cells, or fibrous tissue were trimmed if necessary.
Figure 1: ROIs selected for image analysis a) Hematoxylin-eosin-stained section of sample 22; b,c) Five spots within a tumor were selected, and a 174.4-µm × 130.8-µm field was defined as an ROI for each spot; d) In each ROI, fields without tumor cells, such as stromal cells, or fibrous tissue were trimmed if necessary.
ROI: region of interest.
View Figure 1
All analyses were performed using JMP Pro statistical software (ver. 12.2.0, SAS Institute, Japan). A Wilcoxon, signed rank test was employed to examine for differences in conventional HER2 status and PID counts per cell. A non-parametric Spearman rank order correlation was used to determine the relationship between IHC score and ISH ratio, and between ISH ratio and PID counts per cell. Receiver Operating Characteristic (ROC) curves were generated to evaluate the ability of IHC-PID to discriminate HER2 positive cases versus HER2 negative cases.
All the samples and all 5 spots could be evaluated with IHC-PID. In each spot, approximately 100 to 300 cells were counted.
Representative results of the IHC-PID assays are shown in Figure 2. The exposure time of the bright fields were 6.1 to 7.1 ms, and 18.9 to 22.2 ms for dark fields. The images show bright red "spots" resulting from the PIDs through fluorescence microscopy and they are not "dots" or "particles" in a precise sense. The images of red spots were processed using image editing software.
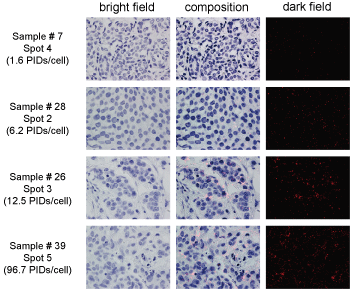 Figure 2: Examples of PID image analysis.
Figure 2: Examples of PID image analysis.
PID images of samples 7, 28, 26, and 39. The PIDs/cell of each section were as follows: Sample 7, spot 4, 1.6 PIDs/cell; sample 28, spot 2, 6.2 PIDs/cell; sample 26, spot 3, 12.5 PIDs/cell; and sample 39, spot 5, 96.7 PIDs/cell. The images shown in the left and right rows were acquired using bright-field or dark-field fluorescence microscopy, respectively. The images in the middle row show the composition of tumor cells and fluorescent spots.
The images show bright red "spots" resulting from the PIDs through fluorescence microscopy and they are not "dots" or "particles" in a precise sense.
PID: phosphor-integrated dot.
View Figure 2
HER2 scores determined using conventional IHC scores, ISH ratios, and PIDs/cell are shown in Table 2. In some cases, HER2 status changes depending on the way of determination. Cases 32 and 33 were determined as HER2 negative according to ASCO/CAP guideline 2007 [9] or determined only with ISH, but as HER2 positive according to ASCO/CAP guideline 2013 [2]. Case 36 was HER2 positive with IHC but determined as negative with ISH. In total, there were 26 HER2 negative and 15 HER2 positive cases according to ASCO/CAP 2013, 28 HER2 negative and 13 HER2 positive cases according to ASCO/CAP 2007, and 29 HER2 negative and 12 HER2 positive cases only with ISH.
Among cases diagnosed as HER2 positive after surgery according to ASCO/CAP guideline 2007, 7 cases received anti-HER2 therapy. Six cases were not treated with anti-HER2 antibody because of complication or financial problem. Two cases who received anti-HER2 therapy experienced recurrence within three years after surgery (Table 2).
Table 3 presents the relationship of IHC (determined according to ASCO/CAP 2013 or 2007) versus ISH (HER2/CEP17 ratio < 2.0: negative; ratio ≥ 2.0: positive). Between IHC score with ASCO/CAP 2013 and ISH, the coefficient of correlation is 0.5236 and p value is 0.0004, while between IHC score with ASCO/CAP 2007 and ISH, the coefficient of correlation is 0.5102 and p value is 0.0007 (Spearman rank order correlation test).
Table 3: Relationship of IHC (determined according to ASCO/CAP 2013 or 2007) versus ISH (HER2/CEP17 ratio < 2.0: negative; ratio ≥ 2.0: positive). View Table 3
Independent of the assay HER2-negative samples had low PIDs/cell (ASCO/CAP 2013, range: 0.6-14.3, median: 2.85; ASCO/CAP 2007, range: 0.6-14.3, median: 2.85; ISH only, range: 0.6-14.3, median: 2.9) and HER2-positive samples had relatively high numbers of PIDs/cell (ASCO/CAP 2013, range: 1.2-153.7, median: 5.3; ASCO/CAP 2007, range: 1.6-153.7, median: 6.8; ISH only, range: 1.6-153.7, median: 11.05) (Figure 3). Conventionally HER2 positive cases showed significantly higher PIDs per cell than those of conventionally HER2 negative cases (P < 0.05 by Wilcoxon signed rank test), regardless of the way of determining HER2 status. There was a significant association between the ISH ratio and PIDs/cell (Spearman rank correlation r = 0.4833, P = 0.0014). Apart from HER2-positive samples (IHC 3+ and/or ISH ratio > 2.0 cases), and in many of the samples, PIDs were homogeneously distributed. However, there were large variations in PID counts among the observation spots (e.g. samples 8, 11, 19, 23, and 26) (Table 2).
 Figure 3: Comparison of PIDs/cell between HER2-negative and HER2-positive tumor samples determined using conventional assays.
Figure 3: Comparison of PIDs/cell between HER2-negative and HER2-positive tumor samples determined using conventional assays.
PIDs/cell was significantly higher in HER2-positive samples compared with those in HER2-negative samples, independent of the method used to determine HER2 status a) HER2 status was determined according to the latest ASCO/CAP guidelines [2]; b) HER2 status was determined according to the previous ASCO/CAP guidelines [9]; c) HER2 status was determined using only ISH (A tumor was considered HER2-positive with an HER2/CEP17 ratio ≥ 2.0 or average HER2 copy number ≥ 6.0 signals/cell, counting ≥ 20 cells within the area. A tumor was considered HER2-negative with an HER2/CEP17 ratio < 2.0 and an average HER2 copy number < 4.0 signals/cell).
IHC: immunohistochemistry; ISH: in situ hybridization; ASCO/CAP: American Society of Clinical Oncology/College of American Pathologists; PID: phosphor-integrated dot, Error bars, standard deviation.
Statistical comparisons were performed using Wilcoxon signed rank test.
View Figure 3
According to the ROC curve analysis, when HER2 expression was determined using the ASCO/CAP 2013 criteria [2], a cut-off value = 3.6 PIDs/cell was determined (sensitivity and specificity, 80% and 62%, respectively, P = 0.0006), and the area under the curve (AUC) was 0.744. When criteria according to ASCO/CAP 2007 [9] were used, the cut-off value was 15.3 PIDs/cell (sensitivity and specificity, 46% and 100%, respectively, P = 0.0002), and the AUC was 0.812. If HER2 status was determined using only the ISH (ratio ≥ 2.0, positive; ratio < 2.0, negative), the cut-off value = 15.3 PIDs/cell (sensitivity and specificity, 50% and 100%, respectively, P = 0.0008), and the AUC = 0.812 (Figure 4).
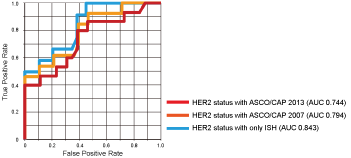 Figure 4: Diagnostic potential of PID nanoparticles.
Figure 4: Diagnostic potential of PID nanoparticles.
Receiver operating characteristic (ROC) curve analyses show that the IHC-PID assay exhibited a significant ability to predict HER2 expression in breast cancer.
Red line: HER2 status was determined using the latest ASCO/CAP guidelines [2]; Orange line: HER2 status was determined using the previous ASCO/CAP guidelines [9]; Blue line: HER2 status was determined using only ISH (A tumor was considered HER2-positive with an HER2/CEP17 ratio ≥ 2.0 or an average HER2 copy number ≥ 6.0 signals/cell, counting ≥ 20 cells within the area. A tumor was considered HER2-negative with an HER2/CEP17 ratio < 2.0 and an average HER2 copy number < 4.0 signals/cell).
Asterisks are put by the curves to point where the cut-offs are located, and the meaning of the color of the asterisks is the same as that of the lines.
IHC: immunohistochemistry; ISH: in situ hybridization; ASCO/CAP: American Society of Clinical Oncology/College of American Pathologists; PID: phosphor-integrated dot; AUC: area under curve.
View Figure 4
PID nanoparticle analysis detected significantly higher levels of HER2 expression in conventionally determined HER2-positive breast cancer samples compared with HER2-negative samples. HER2 status is currently judged visually by expert pathologists. This task is subjective, difficult, and time-consuming. The present data support the conclusion that our novel IHC-PID assay achieves better objective quantitation of HER2 levels compared with standard assays. The intensity of IHC staining depends on the enzymatic activity of streptavidin-conjugated horseradish peroxidase (HRP), which reacts with secondary antibody, and is significantly influenced by the reaction time, temperature and HRP substrate concentration. Therefore, even with using commercial companion software, the quantitative sensitivity of IHC is low. Owing to the high reactivity of PID to secondary antibody, IHC-PID gives us quantitative data with high accuracy and sensitivity [6].
Although the PIDs/cell differed significantly difference between HER2-positive and HER2-negative samples, there was only a modest correlation between the ISH and IHC-PID data (Spearman rank correlation r = 0.4833). But still, this correlation is only slightly lower than that of the correlation between conventional IHC and ISH (r = 0.5236: between ASCO/CAP 2013 and ISH; 0.5102: between ASCO/CAP 2007 and ISH), the results of which were very similar to the previous report [10]. This finding indicates that HER2 expression and HER2 amplification are directly, but not always, concordant [11].
The AUCs determined using ROC curve analyses were high, independent of the type of conventional determination of HER2 status, although the cut-off values varied from 3.6 PIDs/cell to 15.3 PIDs/cell. As shown in Figure 5, all HER2 negative cases showed PIDs/cell less than 15.3 regardless of the HER2 status definition way, and this means that IHC-PID can specifically detect HER2 positive cases if PIDs/cell is higher than 15.3. The problem is the relatively low sensitivity, and the reason of this is that there are some ISH positive cases with very low PIDs/cell, especially HER2 2+ cases, such as case 28 and 35 in Table 2. The large variation of PIDs/cell among HER2 positive cases leads to the large variation of cut-off values from 3.6 up to 15.3. About case 28, ISH ratio was 2.17, which was once determined as "equivocal" (ISH ratio between 1.8 and 2.2) in ASCO/CAP guideline 2007 [9], so HER2 gene may be only slightly amplified in this case. ISH ratio of case 35 was relatively high, but in fact HER2 gene amplified spot is very limited, and not in most of the area. In these cases, HER2 gene may be amplified, but might not be enough for high expression of HER2 protein on cell membranes. Incidentally, these two cases did not receive anti-HER2 therapy after surgery, but do not show clear evidence of recurrence in these seven years. Although these cases are determined as "HER2 positive" in the current guideline but they might not be real candidates of anti-HER2 therapy.
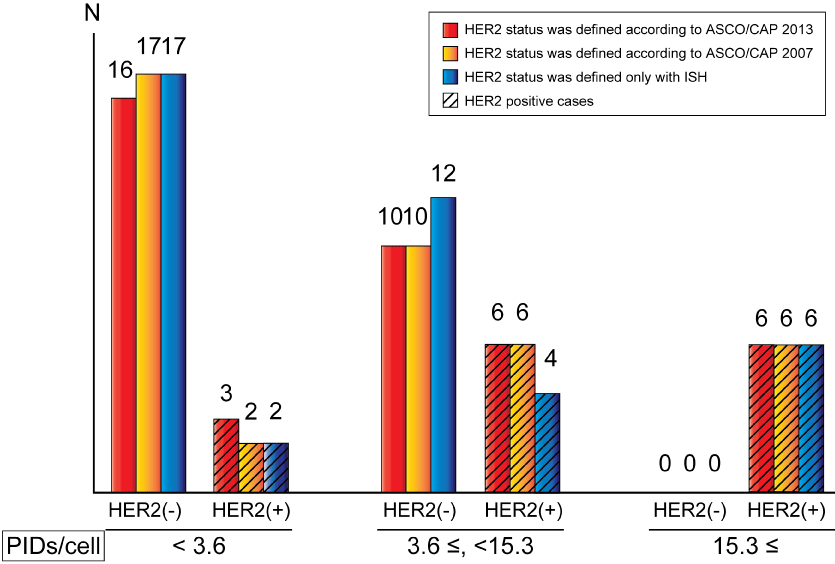 Figure 5: Relationship between PIDs/cell and HER2 status.
Figure 5: Relationship between PIDs/cell and HER2 status.
Cases are divided into three types: Cases with PIDs/cell < 3.6; with 3.6 ≤ PIDs/cell < 15.3; with PIDs/cell ≥ 15.3.
Red bars: HER2 status was determined using the latest ASCO/CAP guidelines [2]; Orange bars: HER2 status was determined using the previous ASCO/CAP guidelines [9]; Blue bars: HER2 status was determined using only ISH (A tumor was considered HER2-positive with an HER2/CEP17 ratio ≥ 2.0 or an average HER2 copy number ≥ 6.0 signals/cell, counting ≥ 20 cells within the area. A tumor was considered HER2-negative with an HER2/CEP17 ratio < 2.0 and an average HER2 copy number < 4.0 signals/cell); Bars applied with oblique lines: HER2 positive cases.
View Figure 5
We found that certain samples varied greatly in the PIDs/cell among the five spots selected. The variation was very high especially among HER3+ and ISH positive cases (such as case 34 and 37-41 in Table 2). However, case 8, which was diagnosed as IHC 1+ and DISH negative, showed a large variation of PIDs/cell among five spots. Based on this result, we reexamined the IHC and DISH slides. We found only a few intense staining of HER2 on IHC slide, and such area was less than 10% of the lesion, so the IHC score is still "negative" on the basis of ASCO/CAP guideline 2013 [2]. But surprisingly, DISH ratio in spot 1 and 2 were 9.23 and 6.24 respectively, while HER2 gene was not amplified in other spots. Case 8 seems to be a real genetical heterogeneous breast cancer, and IHC-PID helped us to find it. In contrast, the PIDs/cell of samples 22, 31, and 35 with heterogeneous IHC staining patters were almost evenly distributed among the five spots (Table 2). These findings may require us to review the level of heterogeneity of HER2 expression in primary breast cancers.
In our procedure, all specimens were fixed in 10% formalin and embedded in paraffin, and this method follows the standard procedures of IHC. Gonda, et al. reported that the actual number of HER2 proteins of formalin-fixed SK-BR3 cells recognized by the anti-HER2 antibody was approximately 5.0 × 104, which was less than the living SK-BR37's HER2 protein number known as 2.0 × 106 per cell. They suggested that this result might due to the effect of formalin fixation [6]. If we use fresher specimens such as live animals, the result of PID examination might show higher PID counts per cell compare to that with formalin-fixed specimen. We need to research farther to investigate the best condition of specimens for the examination with PIDs.
Although IHC-PID is a very promising diagnostic tool in that it gives us quantitative data of protein expression with high accuracy, sensitivity and objectivity, there are some advantages of conventional tool. For example, we can observe the protein expression of a whole tumor at a glance on IHC slides. In IHC-PID, we can assess the expression of small spots. Immunohistochemical procedure of IHC-PID is similar to that of conventional IHC, but image analysis with fluorescent microscope or software is time consuming. Preservation of huge electric data from IHC-PID analysis is also challenging.
Another problem of IHC-PID to be overcome before applying it for clinical use is the high cost. However, this examination is expensive because currently we have to outsource this work, from staining to analysis. If we are well prepared to do the whole process, or even some part of it, in our own facilities, the running cost will be much lower than at present.
Because IHC-PID is a newly developed method, only a few articles are reported at this time. Gonda, et al. first reported the usability of PIDs to detect HER2 protein in breast cancer cell lines and clinical biopsy tissue [6]. Yamaki, et al. detected PD-L1 expression in pancreatic cancer using IHC-PID [7]. As IHC-PID is a promising examination tool, more research for detecting other proteins in other tissues or organs using PIDs is awaited.
The relationship of PIDs/cell and response to anti-HER2 therapy a topic of great interest, however, because of the small cohort and short observation time, we cannot say much about it this time. Further study is essential for us.
In summary, we used a PID nanoparticle assay to analyze HER2 expression in FFPE tissue samples of 41 patients with primary breast cancer. Our results were highly concordant with the conventional assays of HER2 status. Quantitation of HER2 expression using PID nanoparticles may represent a valuable new approach for preclinical and clinical characterization of breast cancer. A larger scale study is required to determine whether the PID nanoparticle assay has predictive and prognostic value for patients with HER2-positive breast cancer patients who are treated with anti-HER2 antibodies.
We thank New Histo. Science Laboratory Co., Ltd. (Tokyo, Japan) for performing IHC with PID nanoparticles. We thank Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
No conflict of interests declared.
The study was carried out with permission from The University of Tokyo Hospital Ethics Committee. Written informed consent was obtained from all subjects.