Breast cancer (BC), a malignant proliferation of the epithelial cells that line the ducts or lobules of the breast, is the most common malignancy in women, accounting for approximately one-third of all cancers in women worldwide. According to its death rate, it is the second most frequent cancer causing mortality for women worldwide. Polymorphism involves one of two or more variants of a particular DNA sequence. The most common type of polymorphism involves variation at a single base pair. In recent years there has been an enormous effort by to identify genetic mutations that play a major role in genetic predisposition to and disease progression in particular cancers. This approach has met with considerable success, as exemplified by numerous studies in cancers. In this review we summarize the correlation between genetic polymorphisms and susceptibility to breast cancer.
Breast cancer, gene polymorphism
Breast cancer is the most common cancer diagnosed in women, accounting for more than 1 in 10 new cancer diagnoses each year. It is the second most common cause of death from cancer among women in the world. Breast cancer always evolves silently. Breast cancer, a highly penetrant hereditary disorder, is the most common cancer in women worldwide. Approximately 10% of breast cancer cases are hereditary and 15% of patients with invasive breast cancer have a first-degree relative with the same disorder. Genetic counseling has become an important tool of the health care system providing information and support to families at risk of a genetic disorder. Most of the patients discover their disease during their routine screening. Others may present with an accidentally discovered breast lump, change of breast shape or size, or nipple discharge. However, mastalgia is not uncommon. Physical examination, imaging, especially mammography, and tissue biopsy must be done to diagnose breast cancer. The survival rate improves with early diagnosis. The tumor tends to spread lymphatically and hematologically, leading to distant metastasis and poor prognosis. This explains and emphasizes the importance of breast cancer screening programs. Breast cancer can be invasive or non-invasive according to its relation to the basement membrane. Noninvasive neoplasms of the breast are broadly divided into two major types, lobular carcinoma in situ (LCIS) and ductal carcinoma in situ (DCIS). LCIS is regarded as a risk factor for the development of breast cancer. LCIS is recognized by its conformity to the outline of the normal lobule, with expanded and filled acini. DCIS is more morphologically heterogeneous than LCIS, and pathologists recognize four broad types of DCIS: Papillary, cribriform, solid, and comedo. DCIS is recognized as discrete spaces filled with malignant cells, usually with a recognizable basal cell layer composed of presumably normal myoepithelial cells. The papillary and cribriform types of DCIS are generally lower grade lesions and may take longer to transform into invasive cancer [1,2] (Figure 1).
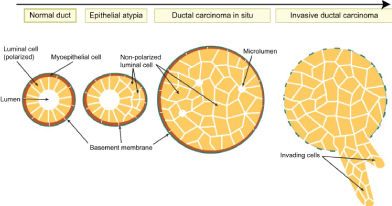 Figure 1: Development of breast cancer.
View Figure 1
Figure 1: Development of breast cancer.
View Figure 1
Genetic polymorphism is a difference in DNA sequence among individuals, groups, or populations. Sources include single nucleotide polymorphisms (SNPs), sequence repeats, insertions, deletions, and recombinations (e.g. a genetic polymorphism might give rise to blue eyes vs. brown eyes, or straight hair vs. curly hair). Genetic polymorphisms may be the result of chance processes, or may have been induced by external agents such as viruses or radiation. If a difference in DNA sequence among individuals has been shown to be associated with disease, it will usually be called a genetic mutation. Changes in DNA sequence that have been confirmed to be caused by external agents are also generally called 'mutations' rather than 'polymorphisms'. Most common polymorphisms are potential regulatory polymorphisms located in noncoding regions, including promoter/upstream, downstream and intron regions that may affect transcription [3] (Figure 2).
 Figure 2: TNM classification of breast cancer.
View Figure 2
Figure 2: TNM classification of breast cancer.
View Figure 2
Tissue inhibitor of metalloproteinase-2 (TIMP-2) is an endogenous inhibitor of MMP-2 that has been implicated in the regulation of MMP-2 proteolytic activity through formation of a 1:1 stoichiometric inhibitory complex with the enzyme [4]. Genetic polymorphisms in the TIMP-2 gene, located on chromosome 17q25, may lead to an increase or decrease in TIMP-2 activity and subsequently disrupt the balance between the activity of TIMP-2 and MMP-2. This disrupted balance could then influence cancer development and progression [5]. TIMP-2 is expected to serve as an anti-invasive/anti-metastatic agent through inhibition of MMP-2. Changes in the level of TIMP-2 are known to directly affect the activity level of MMP-2 [6]. One research explore the association between TIMP-2 and breast cancer, and found that TIMP-2 rs7501477 and rs8064344 mutation affects the genetic susceptibility of breast cancer; while, no effect of rs2277698 mutation on breast cancer was found [7]. A previous study suggested that mutations in the rs4789936 locus not only affect the genetic susceptibility of breast cancer, but also affect the survival of breast cancer patients [7]. So, the role of rs4789936 mutation on the genetic susceptibility of breast cancer remains controversial.
The prognosis of breast cancer patients depends on the clinical or pathological stage at diagnosis. Thus, individuals with hereditary breast cancer could benefit from epigenetic screening for early diagnosis and treatment that prevents the disease from developing. RETN polymorphisms have been identified in various cancers, including colon and lung [8-10]. SNP rs3219175 and rs7408174 at greater risk of developing RA disease [11], and the effects of these variants on resist in expression require to be further examined in breast cancer cells in the future. A previous study has reported that the rs1862513 SNP in RETN increased breast cancer risk and a tendency for luminal breast tumors in Mexican women, which divided into different subtypes according to BMI [12].
The X-ray repair cross-complementing group 3 genes (XRCC3) belongs to a family of genes responsible for homologous recombination, repairing DNA double-strand breaks (DSBs) and interstrand cross-links caused by normal metabolic processes and exposure to ionizing radiation [13]. The XRCC3 gene is structurally and functionally related to the RAD51 gene, which is known to play an important role in all three stages of homologous recombination and catalyzes the invasion of broken ends of the DSB into the intact sister chromatid. Moreover, XRCC3 takes part in DSB repair as it causes slowing of DNA synthesis and recruitment of RAD51 at repair sites [14]. Studies that revealed that polymorphisms in the XRCC3 (rs861539) gene were significantly associated with an increased risk of BC were conducted in different ethnic populations, and the results are inconsistent with our findings, which may be due to environmental factors, ethno-cultural variations, and/or variations in linkage disequilibrium of the XRCC3 gene [15-17].
DNA repair is a part of the barriers, protecting from cancer forming mutations [18,19]. Following the results of studies, cancer diseases are driven by a compromised ability of DNA repair. Therefore, a set of alleles of repair protein encoding genes may largely define the individual abilities for DNA damage repair, as well as the susceptibility to tumor development. It is then important to learn the polymorphic variants of the genes, associated with DNA repairs, as well as with their degradation in their population. The single nucleotide polymorphisms (SNPs) may change the risk of cancer. SNPs may then be regarded as potential markers of carcinogenesis [20,21]. The Asp allele of Gly322Asp polymorphism was strongly correlated with breast cancer. The Gly322Asp polymorphism affected the tumor grade and size, as well as was associated with the lymph node status. As literature data demonstrate DNA damages to be highly significant in the pathogenesis of breast cancer, especially those which require repair by homologous recombination [22,23].
FSCN1, a 55-kDa cytoskeletal actin-binding protein that packages actin filaments into tertiary structures, including microspikes, stress fibers and membrane ruffles, within dynamic cellular structures, enhances cell motility, migration and adhesion. Upregulation of FSCN1 has been indicated in various cancer cell types such as stomach, colon, lung, ovary and breast [24,25]. Fascin expression plays a central role in regulating breast cancer cell morphology, migration and invasion potential [24]. Inhibition of FSCN1 reduces cancer migration and tumor metastasis in prostate and oral squamous cancer cells [26,27]. Furthermore, docosahexaenoic acid reduces FSCN1-dependent breast cancer metastasis [25]. These results suggest that knockdown FSCN1 might be a valuable therapeutic strategy for breast cancer. The human tumor suppressor gene p53 encodes a transcription factor, playing a central role in maintaining cellular integrity by the inhibition of cell growth and stimulation of apoptosis in response to DNA damage. Its role in cancer development is widely studied and well documented [28]. A common single nucleotide polymorphism (SNP) is known in the second position of codon 72 at exon 4 (rs1042522 - CCC to CGC), resulting in a substitution of proline (Pro) to arginine (Arg) in the Pro-rich region [29]. The meta-analysis conducted by Gonçalves, et al. [30] showed that breast cancer patients and control subjects were mainly heterozygous in the Asian population (50.1 and 48.0%, respectively) and the African population (43.9 and 49.7%, respectively), whereas the homozygote Arg/Arg was predominant in America (53.6 and 54.5%, respectively) and Europe (54.1 and 53.4%, respectively). However, the Pro/Arg heterozygous genotype was found to be higher in a Saudi population among healthy women (60.19%) than in women with breast cancer (25%) [31], while the Pro/Arg genotype has been reported to be associated with breast cancer risk in an Iranian population, with 75.55% in breast cancer cases versus 62% in controls [32].
BRCA1 and BRCA2 are structurally similar and of similar size (100 and 70 Kb, respectively). The two genes are considered to be tumor suppressor genes. BRCA1 is located on chromosome 17 at the q21 position and consists of 22 exons (60% of the gene corresponding to exon 11) that encode a protein of 1,863 amino acids. Due to its large size, exon 11 is the main target for mutation detection. BRCA2 is located on chromosome 13 at the 12q position and consists of 27 exons that encode a protein of 3,418 amino acids [33,34]. It is extremely important to perform genetic analysis for BRCA mutations in patients at a high risk of developing breast cancer in Argentina to aid health professionals in addressing the prevention, treatment and prognosis of the disease [35,36]. A recent study has yielded encouraging results on the development of a test gene expression profile from the peripheral blood for the early detection of breast cancer with a prediction accuracy of 79.5%, sensitivity of 80.6% and specificity of 78.3% [37]. The -26 G>A polymorphism in the 5' UTR of BRCA2 has been analyzed in some cancer types. Another study showed a monotonic increase in the frequency of the heterozygous G/A genotype from the lowest to highest risk groups of esophageal squamous cell carcinoma from northern China [38].
The author would like to thank Professor/Hafez Ahmed, professor of Biochemistry, Dubai Medical College for his support.