Early serological diagnosis of trichinellosis is affected by a long-term immunological "silent" period following infection. This emphasises the need for the development of sensitive diagnostic methods to be used when antibodies cannot be detected. In this study, we assessed the usefulness of three DNA sequences as a direct diagnosis method to detect early infection with Trichinella spiralis in peripheral blood by SYBR green real-time PCR using a murine model. Primers were designed from a nuclear repetitive DNA element (Rep), the nuclear ribosomal internal transcribed spacer 2 (ITS2) region, and the mitochondrial large subunit of the ribosomal RNA gene (LSU). CF-1 mice were orally inoculated with 500 muscle larvae of T. spiralis and molecular detection was assessed in blood samples between 1 and 20 days post-inoculation (pi). In addition, antibody detection was evaluated every 5 days by ELISA using excretory-secretory antigens. Results showed that the Rep primers were found to amplify between 5 and 19 days pi and the ITS2 and LSU primers between 6 and 15 days pi, whereas the antibodies were detected at 30 days pi. Therefore, this molecular system could be a useful tool for the detection of early T. spiralis infection, in particularly by using the repetitive DNA element.
Trichinella spiralis, Mice, Early detection, Real-time PCR
Trichinellosis is a parasitic zoonosis with high public health impact caused by the consumption of meat and other products from domestic and wild animals infected with Trichinella spp. The genus Trichinella includes nine encapsulated members (i.e. T. spiralis, T. nativa, T. britovi, T. murrelli, Trichinella T6, T. nelsoni, Trichinella T8, Trichinella T9 and T. patagoniensis) and three non-encapsulated species (i.e. T. pseudospiralis, T. papuae and T. zimbabwensis) [1]. In Argentina, T. spiralis is the etiological agent of Trichinella infection in humans and pigs [2-4], however in the last decade three additional species have been reported, T. britovi, T. pseudospiralis and T. patagoniensis [5-7].
The laboratory diagnosis of human trichinellosis generally is performed by an indirect method based on serological detection of antibodies against parasite's excretion-secretion antigens by enzyme-linked immunosorbent assay (ELISA). However, its early serodiagnosis in both human and animals is hindered by a long-term immunological "silent" period following infection, leading to false negative results [8]. Moreover, both human and animal hosts develop an antibody response that remains detectable for a long time after the acute phase of the disease [9-11] making it difficult to distinguish between recent and past infections.
The serological window period coincides with the beginning of the parenteral phase of the parasite's life cycle, when the newborn larvae (NBL) of Trichinella spp. are released into the circulatory system. The fact that early treatment of the infection may help prevent the establishment of the parasites in the skeletal muscles has driven the search for the detection of DNA from NBL in peripheral blood. The Polymerase Chain Reaction (PCR) showed a high sensitivity in the detection of Trichinella in meat, faeces and blood ([12-16]. The assays accomplished using conventional PCR amplification of a 1.6-kb repetitive sequence element [17] in blood from experimentally inoculated mice showed amplification signal from 5 to 14 days post-inoculation (pi) [14], from 3 to 17 days pi [15] and from 5 to 15 days pi [16]. The studies mentioned above were performed by detecting the Rep marker by conventional PCR. In the last decade, the detection of parasite DNA in the blood has been enhanced with the development of different real-time PCR systems [18-20]. Real-time PCR has several advantages over conventional PCR and includes speed, simplicity, reproducibility and quantitative capacity [21]. One approach for real-time PCR detection is to use fluorescent DNA intercalating dyes, such as SYBR-Green I, which is relatively cost benefit and easy to use [22]. This raises the interesting question of whether the direct detection of early T. spiralis infection can be improved by using highly sensitive and accurate methods such as real-time PCR and primers derived from DNA sequences in multiple copies.
The aim of this study was to assess and compare the usefulness of one mitochondrial and two nuclear DNA sequences (one of which is the repetitive sequence element) for detecting newborn larvae of T. spiralis circulating in the peripheral blood stream of experimentally infected mice that are in the serological window period by SYBR Green real-time PCR.
The reference isolate of T. spiralis (code ISS643), which was obtained from Chile and deposited at the International Trichinella Reference Centre (Rome, Italy), (https://trichinella.iss.it) was maintained by serial passages in CF-1 mice in our laboratory. To test the PCR amplification we used isolates of T. spiralis larvae found in the muscles of pigs from geographically distant locations in Argentina: Senillosa, Province of Neuquén (D216, 68°25´W, 39°0´S); Chacharramendi, Province of La Pampa (D223, 65°38´W, 37°19´S); General Lavalle, Province of Buenos Aires (D237, 56°56´W, 36°24´S); and Maipú, Province of Mendoza (D249, 68°47´W, 32°59´S). All muscle larvae were recovered by artificial digestion according to a published protocol [23]. The isolates were identified as T. spiralis by multiplex PCR assay from ribosomal DNA sequences [24].
Mice used in this study were outbred thirty-day-old CF-1 males weighing 25-30 g. A total of thirty mice were assigned randomly to six groups of five animals each. The groups were housed in separate cages in a room with a 12-h light/dark cycle and given free access to food and water. In each group, four mice were inoculated per os with 500 muscle larvae of T. spiralis (ISS643) and one served as negative control. Five groups were used for the molecular detection of T. spiralis and monitored till 20 days pi; each day, blood samples were collected from one group only, so that each group was subjected to blood sampling every five days. The sixth group was used for serodiagnosis and monitored till day 35 pi, with blood samples being also collected at 5-day intervals. Mice were euthanised at day 45 pi and the entire carcasses were processed by standard artificial digestion to check for infection and estimate the number of larvae per gram (LPG). All procedures contributing to this work comply with the ethical standards of the relevant national guides on the care and use of laboratory animals.
Primers were designed based on the mitochondrial large subunit of the ribosomal RNA gene (LSU), the nuclear ribosomal internal transcribed spacer 2 (ITS2) region, and a nuclear 1.6-kb repetitive element (Rep) from the T. spiralis DNA sequences available in GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) with accession numbers AY851277, AY851266 and X06625, respectively. In addition, a fourth primer pair from the 18S ribosomal RNA gene of Mus musculus (GenBank accession number K01364) was used as amplification control of the real-time PCR, to exclude false negative results caused by PCR inhibitors. All primer sets were designed using Primer Express Software, v. 2.0 (Applied Biosystems, USA). Amplicon lengths were 141 bp for ITS2 and LSU, 144 bp for Rep and 138 bp for 18S (Table 1).
Table 1: Primers evaluated for the detection of Trichinella infection. View Table 1
For DNA extraction, 75 µl of blood from the tail tip of individual mice were collected in 5 mM EDTA. Samples were kept at -20 ℃ before being used for DNA purification. DNA was extracted with the FlexiGene DNA Kit according to the manufacturer's instructions (Qiagen, USA).
The real-time PCR mixture consisted of 500 nM of each primer (LSU, ITS2, Rep and 18S), 12.5 µL PerfeCTa SYBR Green SuperMix, UNG, Low ROX (Quanta Biosciences, USA), and 5 μL of DNA from blood of mice in a 25-µL final volume. The assay was performed on an ABI 7500 platform (Applied Biosystems, USA) under the following cycling conditions: 45 ℃ for 5 min, initial denaturation at 95 ℃ for 3 min, and 40 cycles of 95 ℃ for 15 s and 60 ℃ for 45 s. A final dissociation curve was performed with temperature gradually rising from 60 ℃ to 95 ℃, to verify the amplified products.
The primers derived from the LSU, ITS2 and Rep sequences were proved to amplify the isolates of T. spiralis larvae found in the muscle of pigs from distant geographical locations of Argentina (i.e. D216, D223, D237, D249) and the reference isolate of T. spiralis (ISS643), which was also used for experimental inoculation of mice as described below. The extraction of nucleic acids from isolated T. spiralis muscle larvae was performed from a pool of ten parasites under conditions previously described [2]. Real-time PCR conditions for this analysis were the same as described above, but with a final concentration of 250 nM of each primer and 1 μl of DNA template obtained from muscle larvae.
Amplification reactions of DNA from mice blood and from muscle larvae were run in duplicate.
Amplification products were generated from LSU, ITS2 and Rep by conventional PCR, using the same primers as for real-time PCR. The reaction mixture contained 1X PCR buffer, 1 mM MgCl2, 40 µM of each dNTP, 1 U of Taq DNA polymerase (Invitrogen, Brazil) and 250 nM of each primer. Amplifications of the LSU, ITS2 and Rep fragments were performed using 2 µl of DNA from muscle larvae of the reference isolate ISS643. Thermal cycling conditions were 94 ℃ for 3 min, followed by 40 cycles of 94 ℃ for 60 s, 60 ℃ for 60 s and 72 ℃ for 60 s, and finally a 10 min extension at 72 ℃. The PCR products were cloned into Topo TA Cloning Kit pCR 2.1-TOPO vector (Invitrogen, Brazil) according to the manufacturer's instructions. The inserts were sequenced in both directions using an ABI 3500 Genetic Analyser (Applied Biosystems).
To determine the analytical sensitivity of the primers sets used to detect Trichinella infection, standard curves were constructed from ten-fold serial dilution series ranging between 1.0 × 106 and 1.0 × 100 copies per reaction from each recombinant plasmid containing the LSU, ITS2 and Rep inserts. The concentration of each plasmid was measured using a spectrophotometer Ultrospec 2100 Pro (Amersham Biosciences, Sweden) and converted to the copy concentration using the following equation:
Each standard dilution was analysed in duplicate by SYBR Green real-time PCR under the same conditions as described for the blood analysis of mice. The cycle threshold (Ct) values were plotted against the logarithm of their initial template copy concentrations. The slope and correlation coefficient (R2) of the standard curve were determined using the ABI 7500 software (Applied Biosystems, USA). Amplification efficiency (E) was calculated from the slope of each standard curve using the formula : .
ELISA test was made on polyvinyl microplates (MaxisorpTM), sensitised with 50 µg of excretory-secretory antigens derived from muscle larvae of T. spiralis (ISS643). Plates were incubated with serum samples (diluted 1: 200) for 30 min at 37 ℃ with PBS/Tween 20 at 0.05% containing 3% albumin, and then were washed three times with PBS/Tween 20 at 0.05%. Specific antibodies were detected using Horseradish Peroxidase Conjugated Goat Anti-Mouse IgG (Invitrogen, USA). Colour reaction was developed by adding a solution containing ortho-phenylenediamine (OPD) and H2O2 in citrate buffer at pH5. Absorbance values were read in an ELISA automated microplate reader (BIO-TEK Instruments, USA) at 405 nm.
When comparing the DNA sequences from the reference isolate of T. spiralis (ISS643) and the GenBank sequences used to design primers, we found no differences for ITS2, a simple nucleotide polymorphism (SNP) for LSU and three single variations for Rep. The LSU fragment had an A/G base transition at nucleotide position 49 and the Rep fragment showed the base substitutions A/C, C/T and T/C at nucleotide positions 34, 83 and 89, respectively (data not shown).
LSU, ITS2 and Rep sequences were submitted to the NCBI database under GenBank accession numbers KX709909, KX709908, KX709907, respectively.
The linear relationship of the standard curves along a 6-log range yielded the following correlation coefficients and efficiency values: R2 = 0.984 and 94% for ITS2, R2 = 0.991 and 86% for LSU, and R2 = 0.996 and 84% for Rep. In all three assays, the lower detection limit was 10 copies of the target fragment (Figure 1).
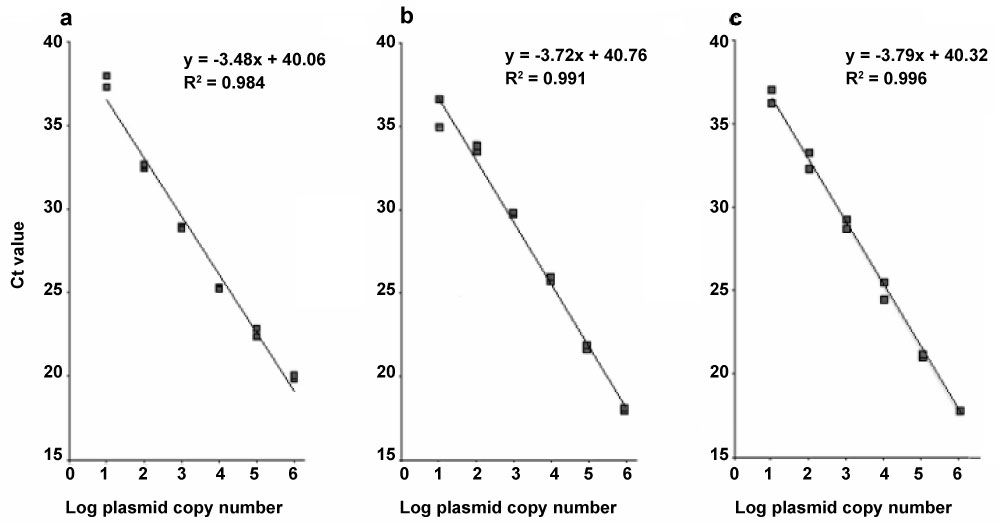 Figure 1: Standard curves for nuclear ribosomal internal transcribed spacer 2 (ITS2), the mitochondrial large subunit of the ribosomal RNA gene (LSU), and the nuclear 1.6-kb repetitive DNA element (Rep) from Trichinella spiralis. Standard curves were generated plotting cycle threshold (Ct) values versus ten-fold serial dilutions (1.0 × 106 to 1.0 × 100 copies µl-1) of plasmid DNA containing cloned target sequences of ITS (a), LSU (b) and Rep (c), amplified with the respective primers using SYBR Green dye assay. All samples were performed in duplicates. View Figure 1
Figure 1: Standard curves for nuclear ribosomal internal transcribed spacer 2 (ITS2), the mitochondrial large subunit of the ribosomal RNA gene (LSU), and the nuclear 1.6-kb repetitive DNA element (Rep) from Trichinella spiralis. Standard curves were generated plotting cycle threshold (Ct) values versus ten-fold serial dilutions (1.0 × 106 to 1.0 × 100 copies µl-1) of plasmid DNA containing cloned target sequences of ITS (a), LSU (b) and Rep (c), amplified with the respective primers using SYBR Green dye assay. All samples were performed in duplicates. View Figure 1
The set of primers derived from the nuclear DNA regions (ITS2 and Rep) and the mitochondrial LSU gene amplified from all five tested T. spiralis isolates, but with different detection levels. The primer Rep exhibited the lowest Ct value (18.18 ± 0.52), whereas those of ITS2 and LSU were 22.40 ± 0.93 and 25.02 ± 0.54, respectively.
All CF-1 mice (n = 24) orally inoculated with 500 muscle larvae of T. spiralis and processed by standard artificial digestion were found infected, with an average parasite burden of 723.87 ± 638.11 LPG. None of the six control animals became infected.
In the mice monitored by real-time PCR for 20 days, the primer Rep was able to detect T. spiralis infection from 5 to 19 dpi. However, detection followed a discontinuous pattern within this period, since the primer yielded negative results at days 9, 10, 17, and 18 pi. Between 25 and 75% of the mice inoculated per cage (n = 4) were found positive for DNA from NBL of T. spiralis (Table 2). Nineteen of the 80 blood samples from the inoculated mice tested (23.75%) were positive, with Ct ranging between 23.71 ± 0.04 on day 7 pi and 39.31 ± 0.47 on day 16 pi (Table 3).
Table 2: Proportion of positive mice relative to the number of experimentally infected mice per cage per sampling day. Trichinella DNA was amplified with the primer sets LSU, ITS2 and Rep by real-time PCR. View Table 2
The primer derived from the ITS2 sequence detected T. spiralis at days 6, 7, 8, 12, 14 and 15 pi, with a proportion of positive animals ranging between 25 and 50% per cage (Table 2). Seven of the 80 blood samples from the inoculated mice tested (8.75%) were positive for T. spiralis. Ct values varied between 26.84 ± 0.37 and 37.12 ± 0.80 at days 7 and 14 pi, respectively (Table 3).
Table 3: Detection of early infection by SYBR green real-time PCR in experimentally infected CF-1 mice. View Table 3
The LSU primer could amplify DNA from NBL of T. spiralis at days 6, 7, 8, 12 and 15 pi and the proportion of positive animals ranged from 25 to 50% per cage. Six of the 80 blood samples from the inoculated mice tested (7.5%) were positive for T. spiralis. Ct values varied between 30.73 ± 0.51 and 33.30 ± 1.61 at days 7 and 12 pi, respectively (Table 3).
No amplification was detected in any of the 20 blood samples from the 5 non-infected mice used as negative control.
The fragment from the 18S region used as positive control for real-time PCR was detected in all blood samples examined (n = 100), showing an average Ct of 22.69 ± 3.58.
The ELISA test revealed an increase in the optical density as a function of time post-inoculation in all four infected animals analysed, which showed a remarkable increase in absorbance from day 30 pi which continued to rise until the end of the experience at day 35 pi, except for one mouse that was only sampled until day 30 pi. The average absorbance difference (ΔAbs) between the first and the last serum sample was 1.12. A maximum ΔAbs of 0.74 was observed between days 0 and 30 pi. Control mouse showed no serological response.
The primers derived from both mitochondrial and nuclear sequences allowed amplification by SYBR Green real-time PCR from the four Argentine isolates of T. spiralis and from the Chilean reference isolate (ISS643). The values of the Threshold Cycle (Ct) are inversely proportional to the amount of target DNA in a sample, and thus a lower Ct value indicates a higher initial amount of target DNA, and vice versa. In the five isolates of T. spiralis, the primers Rep had the lowest Ct value (18.18 ± 0.52), followed by ITS2 (22.40 ± 0.93) and finally by LSU with the highest Ct value (25.02 ± 0.54). The disparity in Ct values among the three primers sets is probably due to different copy numbers of the target DNA sequences, since the three primers pairs had the same detection limit (10 copies), as indicated by the standard curves developed for sensitivity analysis. On the other hand, the similarity in the Ct values observed among the five isolates when they were amplified with the same primer set indicates the ability of the three primer sets to detect the isolates of T. spiralis not only from Chile but also from different regions of Argentina.
All the primers tested on muscle larvae and on NBL circulating in mice blood yielded similar rankings of Ct values, considering the sampling days when the three markers were detected, i.e. at days 6, 7, 8, 12, and 15 pi (Table 3). This ranking of primer sensitivity expressed as Ct values was consistent with the ranking based on the number of blood samples detected positive by each primer set with respect to the total number of blood samples analysed (Rep, 19; ITS2 7; LSU, 6). The primer Rep showed an earlier and longer detection of NBL in mice (from day 5 to 19 pi) than did ITS2 and LSU (both from day 6 to 15 pi) (Table 3). In this paper, the first day of detection of the repetitive DNA element using Rep (day 5 pi) is in agreement with that reported in experimental studies on mice, which evaluated the amplification of this repeated sequence by conventional PCR [14-16], although it was detected earlier (day 3 pi) by using PCR followed by Southern blot analysis [7]. However, to our knowledge, the amplification of the primers Rep until day 19 pi represents the longest period during which acute infection with Trichinella was detected by molecular methods.
The amplification of the three primer pairs was discontinuous, as they were not detected every day within the monitoring period (i.e. Rep, days 5 to 19 pi; ITS and LSU, days 6 to 15 pi) (Table 3). Likewise, a fluctuating pattern of detection of T. spiralis was observed for the group of four inoculated mice per cage, where in each sampling day we found one to three positive mice but not all of them (Table 2). These results are in agreement with other similar studies in mice using conventional PCR [16] and may be explained by the random presence of a NBL or part of it in the blood sample.
In our study, the nucleotidic sequencing of the amplification products from the reference isolate of T. spiralis using the three primers sets revealed a SNP in the LSU fragment and three SNPs in the Rep fragment. Although these variations did not affect the amplification and detection of the analysed sequences by SYBR Green Real-Time PCR, they should be considered when designing fluorescent probes (e.g. TaqMan probes).
The serological analysis by ELISA-IgG using excretory-secretory antigens of T. spiralis in experimentally inoculated mice showed a noticeable increase in antibodies from day 30 pi onwards (Figure 2). Our result is in accordance with other studies reporting a long-term immunological "silent" period following Trichinella infection in animals and humans [8].
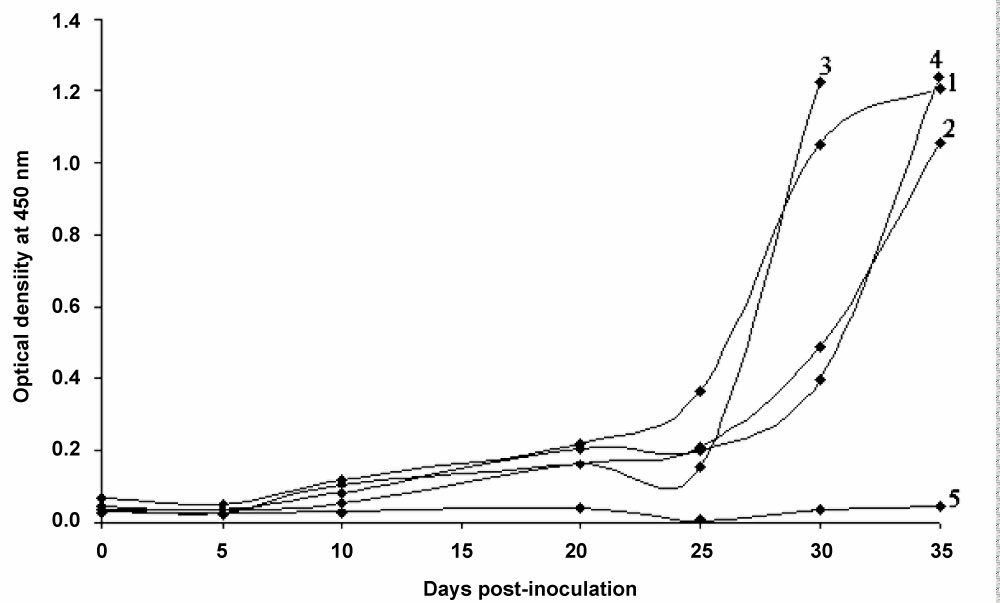 Figure 2: Serological detection of Trichinella spiralis in experimentally infected mice by ELISA.
Figure 2: Serological detection of Trichinella spiralis in experimentally infected mice by ELISA.
Detection curve of circulating anti-Trichinella antibodies by ELISA technique using excretory-secretory antigens from sera of four CF-1 mice (1-4) inoculated orally with 500 muscle larvae of T. spiralis (ISS643) and a non-infected animal (5) used as negative control. All four infected animals showed a remarkable increase in absorbance from day 30 pi which continued to rise until the end of the experience at day 35 pi, except for mouse 3 that was only sampled until day 30 pi. View Figure 2
In conclusion, primers designed in the present study to amplify Rep, ITS2 an LSU genes were efficient to detect T. spiralis during immunological "silent" period (30 days pi) by SYBR Green real-time PCR. Of the three primers sets tested, Rep showed the highest sensitivity for detecting DNA from NBL in the circulating blood of experimentally infected mice from days 5 to 19 pi, as well as for detecting DNA from the muscle larvae of the five geographical isolates. Our results indicate that this molecular detection system for NBL of Trichinella, especially using the repetitive DNA element (Rep), could be a valuable tool during the early stage of the infection when antibodies cannot be detected. In addition, the detection method used in the present study based on SYBR Green real-time PCR does not require a probe, which reduces time consuming and running costs.
We are grateful to Hector F. J. Bergagna, Fernando D. Garofalo, Jorge Gustavo Montali and Roberto H. Paredes for providing parasite material and to Silvia Pietrokovsky for her useful comments on the draft manuscript. This research was supported by the Departamento de Parasitología, Instituto Nacional de Enfermedades Infecciosas, Administración Nacional de Laboratorios e Institutos de Salud, "Dr. Carlos G. Malbrán.