Correct measurements of residual viremia and reservoir size are crucial in HIV-1 eradication trials and there is a need for sensitive and automated assays. The increasing worldwide diversity of HIV-1 subtypes stresses the importance of subtype independent assays.
We added an ultra-centrifugation step to the COBAS AmpliPrep/COBAS TaqMan HIV-1 test, version 2.0 and evaluated assay sensitivity. A previously published method for quantification of total HIV-1 DNA was also evaluated. Nineteen clinical samples from virologically suppressed cART treated patients infected with various HIV-1 subtypes were analysed. The relationship between plasma HIV-1 RNA and HIV-1 DNA in Peripheral Blood Mononuclear Cells (PBMC) was studied.
The HIV-1 RNA PCR assay reached a limit of quantification of 3 copies/ml of plasma. The total HIV-1 DNA assay allowed quantification to 3 copies/million cells with an intra- and inter-assay coefficient of variation of 13.2% and 23.2% respectively. All subtypes studied were quantified by the HIV-1 RNA PCR assay and the HIV-1 DNA assay quantified all subtypes except CRF_AE. No significant correlation was observed between plasma HIV-1 RNA and total HIV-1 DNA in PBMC.
By modification of a commercial assay, we achieved a sensitive quantification of plasma HIV-1 RNA that could be used to assess residual viremia. Sensitive quantification of total HIV-1 DNA in PBMC was also demonstrated. The assays were subtype independent to a large extent, making them useful for research and clinical use in settings where a variety of HIV-1 subtypes is present.
Treatment of HIV-1 infection with effective combination Antiretroviral Treatment (cART) reduces plasma viral load to undetectable levels measured with clinical assays [1,2]. However, very stable viral reservoirs established early in infection are obstacles for eradication [3-5]. Precise measurements of the residual viremia that is released from these reservoirs combined with methods to estimate viral reservoir size are important in research for a cure of HIV-1. In recent years it has also been proposed that at least in some patients it may be clinically relevant to monitor viremia even below 50 HIV-1 RNA copies/ml [6,7]. The commercial assays of today have a limited sensitivity and a high variability in samples < 20 to 40 copies/ml depending on the assay [8].
The Single Copy Assay (SCA), a quantitative assay that can detect less than 1 copy of HIV-1 RNA/ml, has been widely used for research purposes in recent years [9-13]. This assay is labour-intensive and is currently limited to quantification of subtype B strains [14]. The methods need to be sensitive but also subtype-independent since many research and clinical environments are dealing with a diversity of HIV-1 subtypes. Several recent studies have reported increasing HIV-1 subtype diversity in different parts of the world, including Sweden and USA [15-18]. Previous reports have described in-house assays or modified commercial assays that increase their sensitivity [19-21]. For example, Havlir, et al. increased the sensitivity by using ultra-centrifugation prior to amplification with the Roche Amplicor Ultrasensitive assay [19]. Another, more recent study showed a single-copy sensitivity by increasing the sample volume, and adding an ultra-centrifugation step to the Abbott Real Time assay [21]. However, information about subtypes were lacking in these studies. Modified commercial assays have the advantage of wide availability, being extensively tested, standardised and automated. It should also be possible to detect different subtypes with good reproducibility using these assays. The COBAS Taqman HIV-1 test, version 2.0 from Roche (AS-CT2) has a reported lower limit of detection than the Amplicor and Abbott Real Time Assay [22,23], and the advantage of targeting both gag and LTR, which could increase the accuracy of the viral load determination and the spectrum of quantifiable HIV-1 isolates.
A possible strategy for HIV-1 eradication is to reactivate latent virus and thereby accelerate viral clearance in the presence of cART [24-28]. In studies evaluating effects on the latent reservoir it is necessary to use methods that are able to accurately measure the fraction of replication-competent virus in the reservoir. Currently, the Infectious Units per Million Cells (IUPM) assay is considered the most sensitive and precise assay for measuring HIV-1 viral reservoirs [29]. This assay is however labour intensive and it also has a high variance, a feature making it difficult to measure small differences in replication-competent virus with accuracy. Furthermore it seems to underestimate the actual replication-competent viral reservoir [30]. The measurement of total HIV-1 DNA (linear and circular non-integrated and integrated) and proviral HIV-1 DNA (integrated) has been proposed as surrogate measures of the viral reservoir [31-33]. Since linear unintegrated HIV-1 DNA has a short half-life in vitro and circular HIV-1 DNA is considered being produced during active replication, it is reasonable to assume that most of the HIV-1 DNA will be in an integrated form if replication is halted by cART for a long period of time. Measures of total and proviral HIV-1 DNA have therefore been proposed to be interchangeable surrogates for reservoir size [31]. However, these methods still have the disadvantage of measuring also non-viable virus and thereby risking overestimating the actual viable viral reservoir.
In this study, we added an ultra-centrifugation step prior to the routine RT-PCR with AS-CT2, to enable subtype independent quantification of residual viremia. We also evaluated an assay for ultrasensitive measurement of total HIV-1 DNA in Peripheral Blood Mononuclear Cells (PBMC). Clinical performance of both assays was evaluated in a correlation study between total HIV-1 DNA and plasma HIV-1 RNA in patients on long-term effective cART with different HIV-1 subtypes.
All clinical samples were collected from the Department of Infectious Diseases at Sahlgrenska University Hospital in Gothenburg, Sweden. Nineteen patients who had been on cART for more than 1 year and who had suppressed viral load in plasma (< 50 HIV-1 RNA copies/ml for a median of 54 months), were included in the study. All patients but two had undetectable HIV-1 RNA at study entry (measured by AS-CT2). Eleven patients were infected with HIV-1 subtype B. Four patients were infected with subtype C, two patients with subtype CRF-AE, one patient with subtype D, and one was determined as probably subtype F, according to the HIV-POL subtyping algorithm of Stanford (HIV drug resistant database HIVDB.standford.edu) (Table 1). For statistical calculations in the correlation study, detected but not quantifiable samples were denoted as the mean value in the range between 0 copy/ml and the suggested limit of quantification derived from the sensitivity panels for each assay. Negative samples were denoted as 0.1 copies/ml for HIV-1 RNA and 0.1 copies/million cells for HIV-1 DNA.
Table 1: Characteristics of 19 clinical samples. View Table 1
For the low-copy-number HIV-1 RNA panel we used two different plasma samples from individuals with a known high viral load in plasma. For assessment of reproducibility and limit of detection of the real time PCR for quantification of total HIV-1 DNA we used two different samples from individuals with subtype B viral clades with known high viral loads in plasma.
Low-copy-number HIV-1 RNA measurement panels: A plasma sample with viral load of 268,325 HIV-1 RNA copies/ml was diluted in HIV-seronegative plasma to give the estimated HIV-1 RNA concentrations of 2683, 537, 107, 26.8, 5.4 and 2.7 copies/ml (Panel 1). A second plasma sample was treated in the same way to give the estimated HIV-1 RNA concentrations of 27, 9, 3, 1 copies/ml (Panel 2). The initial viral load was determined by standard PCR with AS-CT2 according to the manufacturer. The total SD in log in this range is reported as 0.06 and total log normal CV 15% according to the manufacturer. The dilutions were stored in 50 ml tubes and frozen at -70 ℃.
Each sample in the HIV-1 test panels was analyzed in quadruplicate with the ultrasensitive and the standard method, to compare assay sensitivity.
All subjects were studied under research protocols approved by institutional review board of the Sahlgrenska Academy. Informed verbal consent was obtained from all patients.
Ultra-centrifugation: For the HIV-1 RNA panel, diluted plasma samples (10 ml) were concentrated by ultra-centrifugation at 50,000 × g at 4 ℃ for 60 minutes. The pellet was resuspended in 1 ml of HIV-1 negative plasma resulting in a 10-fold concentration of the original sample. The suspension was vortexed and transferred to PCR tubes, and stored in -70 ℃. No internal standard for the ultra-centrifugation step was used. In the clinical samples, 6 to 9 ml of plasma was used, dependent on the amount of plasma available.
RT-PCR assay: Except for the ultra-centrifugation step, all samples were handled in the same way, and analyzed with AS-CT2 according to the manufacturer's instructions. Limit of quantification for AS-CT2 is reported to be 20 copies/ml and an internal control for the sample preparation procedure and successful PCR amplification was added to every sample (HIV-1 Quantitation Standard).
Realtime PCR for quantification of total HIV-1 DNA: We used a single-step real time PCR to quantify total HIV-1 DNA in 25 μl of PCR reaction mix containing 13 μl of TaqMan Universal PCR Master Mix, 5 μl of chromosomal HIV-1 DNA and primers and probes as described below. We used a previously described protocol, except that we used half the sample size per reaction mix and another set of primers and probes [34,35]. Cycling conditions: 10 min activation step at 95 ℃ followed by 15 s at 95 ℃ and 60 s at 62 ℃ for 45 cycles. We used a primer-probe set for HIV-1 DNA quantification designed to bind to the conserved region of LTR: primers 5'-TTAAGCCTCAATAAAGCTTGCC-3', and 5'-GTTCGGGCGCCACTGCTAGA-3' with probe 5'FAM-CCAGAGTCACACAACAGAGGGGCA-3'TAMRA.
The clinical samples for the correlation study were analyzed at AIDS Research Institute, IrsiCaixa Foundation, Hospital Universario Germans Trias i Pujol, Barcelona, Spain. Sensitivity test and intra/inter-assay runs were performed at the Department of Clinical Virology, Gothenburg, Sweden.
Generation of a total HIV-1 DNA standard: We used a standard curve prepared by amplification of quantities ranging from 10 to 106 copies of plasmid pCR2.1 harbouring LTR-LTR junction and the CCR5 human gene. The plasmid was kindly provided by Dr Mario Stevenson. The following primers and probes were used: primers CCR5 5'-GCTGTGTTTGCGTCTCTCCCAGGA-3', 5'-CTCACAGCCCTGTGCCTCTTCTTC-3', primers LTR 5'-TTAAGCCTCAATAAAGCTTGCC-3', 5'-GTTCGGGCGCCACTGCTAGA-3' and probes CCR5 5'FAM-AGCAGCGGCAGGACCAGCCCCAAG-TAMRA-3', LTR 5'FAM-CCAGAGTCACACAACAGAGGGGCA-3'TAMRA. Reproducibility and limit of detection. To evaluate the reproducibility of the real time PCR assay for quantification of total HIV-1 DNA, we performed intra-assay and inter-assay tests using a clinical sample of known high plasma viral load. We used geometric Coefficient of Variation (CV) to estimate dispersion [36].
The limit of detection was determined by using a clinical sample in a serial dilution with lymphocytes from a healthy donor.
Linearity of HIV-1 RNA quantification by the ultrasensitive assay: The assay accurately quantified a range of HIV-1 RNA concentrations obtained by serial dilution (Figure 1).
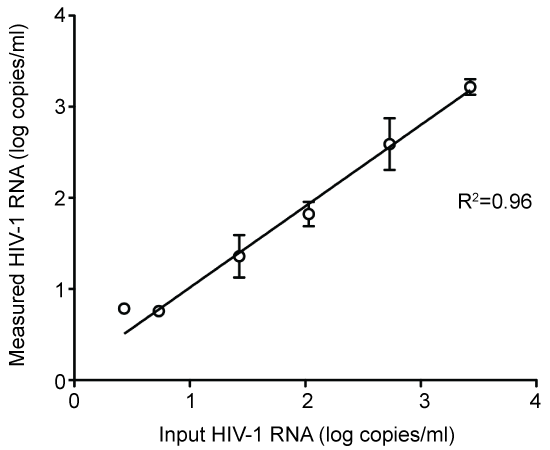 Figure 1: Linearity for measurements with ultrasensitive HIV-1 RNA PCR assay. In-put levels represent theoretical concentrations calculated from dilution of a plasma sample with a HIV-1 RNA level determined by COBAS Ampliprep/COBAS Taqman HIV-1 test, version 2.0. All samples were run in quadruplicate, once in each dilution step. View Figure 1
Figure 1: Linearity for measurements with ultrasensitive HIV-1 RNA PCR assay. In-put levels represent theoretical concentrations calculated from dilution of a plasma sample with a HIV-1 RNA level determined by COBAS Ampliprep/COBAS Taqman HIV-1 test, version 2.0. All samples were run in quadruplicate, once in each dilution step. View Figure 1
HIV-1 RNA sensitivity: The ultrasensitive assay showed greater sensitivity and was able to detect and quantify HIV-1 RNA at lower plasma concentrations than the standard assay (Table 2). In the first panel the ultrasensitive method failed to quantify HIV-1 RNA at 2.7 copies/ml in three out of four samples, but the assay detected HIV-1 RNA in all the wells at this dilution step (Table 2, panel 1). To further evaluate assay sensitivity, a second dilution series with a lower range of estimated copy numbers was created. In this panel, the ultrasensitive assay consistently detected HIV-1 RNA in dilutions estimated to contain 3 copies/ml and 1 copy/ml (in three out of four wells at both dilution steps). The mean value for the obtained concentration was 2.5 copies/ml (SD 0.6 copies/ml) at the 1 copy/ml input level (Table 2, panel 2).
Table 2: Comparison of The COBAS Ampliprep/COBAS Taqman HIV-1 Test v2 (Standard assay) with an ultra-centrifugation step prior to the standard protocol (Ultrasensitive assay) in two separate dilution series (Panel 1 and 2). Input copies/ml are theoretical concentrations calculated from a dilution series originated from a plasma sample quantified by COBAS Ampliprep/COBAS Taqman HIV-1 Test, version 2. All samples were run in quadruplicate (1-4), once in each dilution step. View Table 2
Reproducibility and limit of detection for quantification of total HIV-1 DNA: Figure 2 and Figure 3 show slopes (-3.37 for LTR and -3.34 for CCR5) and linear regression for both LTR and CCR5 genes (R2 = 0.99 respectively) for plasmid pCR2.1.
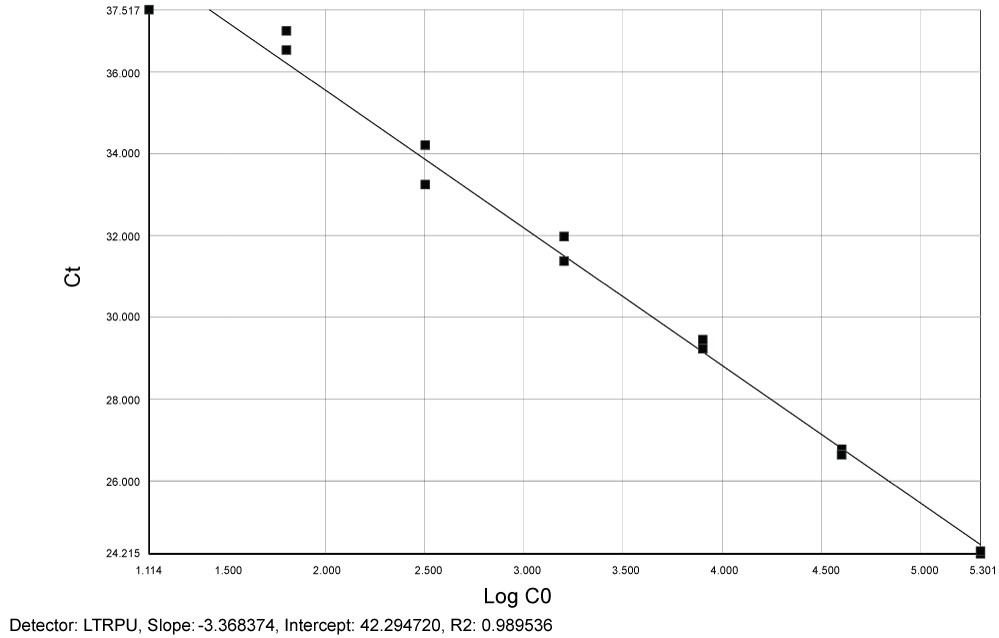 Figure 2: Standard curve generated by plasmid pCR2.1 showing slope (-3.37) and linear regression (R2 = 0.99) for LTR. Ct = Cycle threshold. Log C0 = Log LTR. View Figure 2
Figure 2: Standard curve generated by plasmid pCR2.1 showing slope (-3.37) and linear regression (R2 = 0.99) for LTR. Ct = Cycle threshold. Log C0 = Log LTR. View Figure 2
 Figure 3: Standard curve generated by plasmid pCR2.1 showing slope (-3.34) and linear regression (R2 = 0.99) for human genome CCR5. Ct = Cycle threshold. Log C0 = Log CCR5. View Figure 3
Figure 3: Standard curve generated by plasmid pCR2.1 showing slope (-3.34) and linear regression (R2 = 0.99) for human genome CCR5. Ct = Cycle threshold. Log C0 = Log CCR5. View Figure 3
The geometric CV in five replicates for the intra-assay test was 13.2% and for the inter-assay test the geometric CV in five runs of duplicates was 23.2% (Figure 4). The assay detected and quantified HIV-1 DNA down to the range of 1.4 to 4.6 copies/million cells suggesting the limit of quantification to 3 copies/million cells (Table 3).
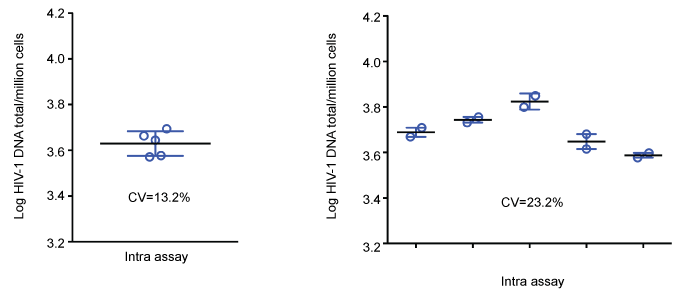 Figure 4: Total HIV-1 DNA PCR intra- and inter-assay validation. Intra-assay: mean log HIV-1 DNA was 3.63/million cells (4291 copies/million cells) in five replicates, SD log HIV-1 DNA 0.054/million cells, geometric CV 13.2%. Inter-assay: mean log HIV-1 DNA in five runs of duplicates was 3.70/million cells (5088 copies/million cells), SD log HIV-1 DNA 0.09/million cells, geometric CV 23.2%. View Figure 4
Figure 4: Total HIV-1 DNA PCR intra- and inter-assay validation. Intra-assay: mean log HIV-1 DNA was 3.63/million cells (4291 copies/million cells) in five replicates, SD log HIV-1 DNA 0.054/million cells, geometric CV 13.2%. Inter-assay: mean log HIV-1 DNA in five runs of duplicates was 3.70/million cells (5088 copies/million cells), SD log HIV-1 DNA 0.09/million cells, geometric CV 23.2%. View Figure 4
Table 3: Dilution series for determining limit of quantification of total HIV-1 DNA PCR assay. View Table 3
Clinical assay performances and correlation of total HIV-1 DNA and plasma HIV-1 RNA: In samples from 19 HIV-1 positive patients measured by AS-CT2, 14 had non-detectable plasma HIV-1 RNA levels, three had detectable but not quantifiable levels and two patients had 24 copies/ml and 45 copies/ml respectively. HIV-1 RNA was detected in 14 out of 19 samples by the ultrasensitive method. In 9 of these 14 positive samples, HIV-1 RNA was quantified in the range of 2.3 to 34.5 copies/ml. The HIV-1 RNA strains detected had previously been identified as subtype B in 9 cases, subtype C in 1, subtype CRF-AE in 2, subtype D in 1, and 1 had been determined as probably subtype F (Table 1).
HIV-1 DNA was detected in 16 out of 19 samples and was quantified at levels ranging from 0.8 to 155.6 copies/million cells. The assay detected ten subtype B strains, four subtype C, one subtype D and one probable subtype F strain (Table 1). HIV-1 DNA was not detected in any of the subtype CRF-AE strains. No correlation was found between plasma HIV-1 RNA and total viral HIV-1 DNA (p = 0.29, r = 0.28) (Figure 5).
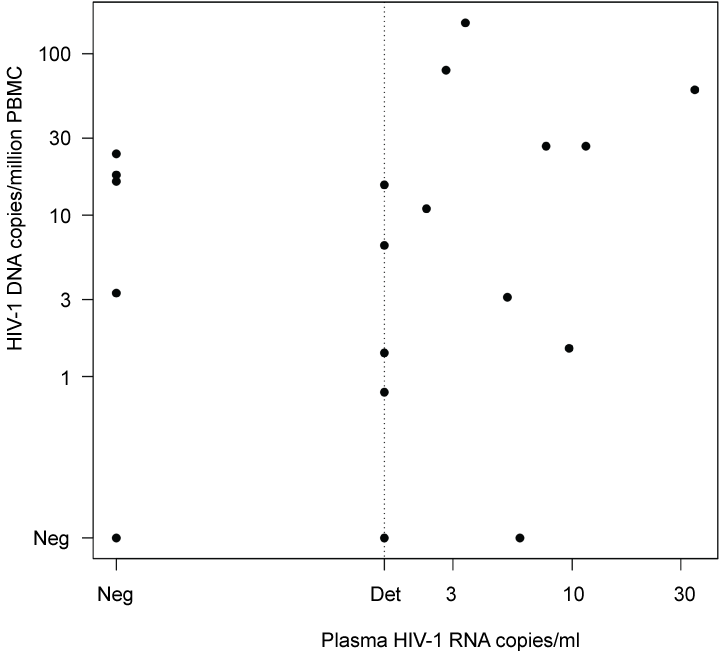 Figure 5: Correlation of total HIV-1 DNA copies/million cells in PBMC and HIV-1 plasma RNA copies/ml in 19 patients with long-term suppressive cART. No significant correlation was seen (p = 0.28, Spearman r = 0.26). Det = detected but not quantified. Neg = negative. View Figure 5
Figure 5: Correlation of total HIV-1 DNA copies/million cells in PBMC and HIV-1 plasma RNA copies/ml in 19 patients with long-term suppressive cART. No significant correlation was seen (p = 0.28, Spearman r = 0.26). Det = detected but not quantified. Neg = negative. View Figure 5
In this study, we present a sensitive and subtype independent quantification of plasma HIV-1 RNA, and a real-time PCR for quantification of total HIV-1 DNA. By adding an ultra-centrifugation step prior to the COBAS Taqman HIV-1 test, version 2.0, we obtained increased sensitivity, quantifying HIV-1 RNA down to 3 copies/ml plasma. In our study we did not attempt to concentrate virus from more than a maximum 10 ml of plasma, but a larger volume might have lowered the limit of quantification in the assay further. There is a considerably large variation when comparing the two sensitivity panels, around 0.5 log. This variation could be expected but would probably be reduced by a larger number of panels.
The previously described single copy assay has the disadvantage of being labour intensive and is developed only for detection of subtype B strains [14]. Other reports have described successful modifications of the Abbot Real Time assay and the Amplicor Ultrasensitive assays, including an ultra-centrifugation step and increased sample volumes [19,21], but did not evaluate amplification of different subtypes. To our knowledge this is the first report of an ultrasensitive detection of various subtypes by a modified commercial HIV-1 RNA assay. The assay showed good clinical performance, detecting residual viremia in all subtypes tested. HIV-1 RNA was not detected by the ultrasensitive assay in five samples, and may indeed reflect true negative samples. It should be pointed out that we used smaller plasma volumes in the clinical performance test (due to limited access) than in the sensitivity panel (6 to 9 ml compared to 10 ml), which has an impact on the limit of detection. Although the number of samples was small, our results indicate that the ultrasensitive assay could be used for measurements of residual viremia in various HIV-1 subtypes.
We also evaluated quantification of total HIV-1 DNA in PBMC by real time PCR and found good reproducibility and sensitivity, with a suggested limit of quantification to 3 copies/million cells and geometric intra-and inter-assay CVs of 13.2% and 23.2% respectively. The assay quantified total HIV-1 DNA in all subtypes tested except CRF_AE. This was probably due to primer mismatch since none of the CRF_AE samples were detected by the HIV-1 DNA assay. One subtype B strain was not detected by the HIV-1 DNA assay, which was probably due to very low viral levels since this sample was also not detected by the HIV-1 RNA assay.
When comparing total HIV-1 DNA in PBMC and plasma HIV-1 RNA in patients on suppressive treatment, no significant correlation was found. A correlation between total HIV-1 DNA in PBMC and plasma residual viremia (HIV-1 RNA) would indicate that a major part of the viremia is derived from resting CD4+ T-cells, and might indicate that a combination of these assays could be used to estimate the reservoir size. One previous study reports a correlation between plasma residual viremia and the frequency of CD4+ T-cells carrying HIV-1 DNA (measured by total HIV-1 DNA). In this study however, nearly 40% of the study participants with undetectable plasma viremia exhibited a relatively high frequency of HIV-1 infection in the CD4+ T-cell compartment. Furthermore, no correlation between activation markers and plasma viremia was found, indicating the possibility of measurement of non-productive virions [37]. However, in a more recent study, Gandhi, et al. showed that residual viremia was associated with IUPM in patients on suppressive cART [38]. Buzon, et al. showed a correlation between the frequency of cells with replication-competent HIV-1, and the total amount of HIV-1 DNA in suppressed patients with treatment initiated at different phase of the infection. However, in this study, residual viremia did not seem to be dependent on the levels of HIV-1 DNA [39].
The lack of correlation in this study could be due to lack of precision and sensitivity of the assays used in the very low levels of HIV-1 RNA and HIV-1 DNA that were present in the tested samples. Residual viremia is usually estimated to around 3 HIV-1 RNA copies/ml and below [13,40] and stochastic influences might have influenced the obtained concentrations. We also had a small sample size, although a larger sample size might not have changed the outcome of this particular result. Alternative explanations may be productive viral reservoirs other than latently infected CD4+ T-cells [41-43]. Low levels of replication and cell-to-cell spread of virus in other compartments may not be reflected in the levels of plasma residual viremia, even as measured by the most sensitive assays.
Recent studies have also questioned the use of PCR assays to determine reservoir size as they do not differentiate between replication-competent proviral HIV-1 DNA and incomplete HIV-1 DNA that cannot produce virions [44]. However, PCR measurements are less costly and labour-intensive to perform than for example IUPM, and there are studies proposing these techniques as surrogate measures of reservoir size [31-33]. Previous studies have suggested that measurement of both total and integrated HIV-1 DNA may be important in estimating reservoir size [32]. In our material, the patients were treated for at least one year prior to sampling and most of the patients were successfully treated for several years. This timeframe should be sufficient for the eradication of other forms of HIV-1 DNA than the integrated pro-viral genome [31]. Again, many of the integrated pro-viruses may not be viable. There are also reports indicating that some patients may have an excess of unintegrated forms of HIV-1 DNA despite long-term cART [45].
This study has some limitations. We did not incorporate an internal standard in the ultrasensitive HIV-1 PCR to monitor the recovery rate of HIV-1 RNA through the ultra-centrifugation and extraction procedures. Furthermore, control copies in the reactions were estimated through PCR, which introduces some uncertainty in the control dilution step. As mentioned above, we also had a limited number of clinical samples tested and future studies, incorporating more samples with diverse subtypes will evaluate this assay further.
In conclusion: by modifications of a commercial assay, ultrasensitive quantification of plasma HIV-1 RNA down to 3 copies/ml was achieved, and quantification of total HIV-1 DNA in PBMC in the range of 3 copies/million cells by real time PCR was documented. The HIV-1 DNA assay need to be modified to incorporate also CRF subtypes and need further testing considering the small sample size. However, the assays were subtype independent to a large extent, making them suitable for research and possibly also clinical use in settings where a variety of HIV-1 subtypes are present.
This work was supported by the Swedish Research Council (K2008-58P-20930-01-4), the Sahlgrenska Academy at the University of Gothenburg (ALFGBG-141741), and Gothenburg Medical Society.
Tomas Mellberg, Jon Krabbe, Maria J. Buzon, Ulrika Noborg, Staffan Lindh, Staffan Nilsson and Bo Svennerholm have all declared that no competing interests exist. Magnus Gisslén has the following potential conflicts: Received honorarium from Abbvie, BMS, Gilead, GSK/ViiV, and Janssen for participation on scientific advisory boards and educational lectures.