Cancer cell presents a dynamic nature that evolves over time in its microenvironment through a complicating molecular and cellular interaction network. Despite the progress already achieved in the imaging technologies and the molecular tools used for tumor cell diagnosis and treatment approaches, metastasis still remains a major hindering factor that limits the clinical outcomes. Circulating tumor cells (CTCs) present the potential in providing critical information to understand the metastasis process, the tumor cell and genome heterogeneity, in a way to overcome cancer therapy bottlenecks and improve the efficacy and safety therapeutic profiles. The experience already gained shows that CTCs as tools possess a predictive biomarker capacity by aiming the understanding of most of the known cancer cell hallmarks at the molecular level, as well as the translation of the extracted relevant knowledge in clinical practice. The efficient isolation and characterization of CTCs might reveal the intratumoral heterogeneity present in tumors while providing critical insights into cancer metastasis. Moreover, by combining the emerging single cell omics technologies, CTCs can be valuable biological source material to unveil the existed intratumoral cellular and genomic heterogeneity, as well as to pharmacologically exploit critical knowledge for the validation of prognostic and diagnostic biomarkers within the concept of precision and personalized cancer therapy. In this article, a novel combined methodology for the isolation and detection of CTCs based on flow cytometry and cellular filtration technologies is presented.
Circulating tumour cells, CTCs, Flow cytometry, Size filtration, Peripheral blood, Liquid biopsies, Novel combined methodology, Isolation and detection of CTCs
CTCs: Circulating tumor cells; EMT: Epithelial mesenchymal transition; PB: Peripheral blood
The microenvironment and epithelial-to-mesenchymal transition (EMT) contribute to intratumoral cellular and genomic heterogeneity by providing neoplastic cells the needed capacity to develop, progress and metastasize [1-3]. Circulating tumor cells (CTCs) are cancer cells derived from primary tumor site that, by crossing the EMT barrier, circulate in plasma and are capable to pass through the endothelial blood vessels thus metastasizing to create secondary tumors [4]. CTCs can be detected very early, before cancer is clinically confirmed and may lead to recurrence or metastatic disease. Crucially, the presence of CTCs constitutes an accessible cellular material with a tumor development monitoring value when metastases are not detected, and histological diagnostic assays are not possible. Clinical studies have confirmed that a numerical increase of CTCs in the patient's bloodstream is an unfavourable prognostic factor in many aggressive tumors like metastatic melanoma, breast and colorectal cancer [5-7]. Furthermore, tumor genetic heterogeneity is observed in CTCs, which include many types of "dormant", apoptotic and aggressive cells and being responsible for therapeutic problems due the change and resistance ability to targeted treatments [8]. According to published data, single cell molecular profiling of CTCs will be essential for potential targeted personalized treatments by interrelating genomic, transcriptomic and proteomic patient's data, providing the opportunity for in vitro functional studies instead of circulated tumor DNA assays (ctDNA), and guiding improved clinical outcomes [9].
A significant number of CTCs selection and analysis approaches are developed in recent years, which shows a rapid development in "non-invasive" methodologies allowing their introduction in cancer prognosis, diagnosis and monitoring therapeutic routine practices and guidelines [10]. Notably, the advanced improvements in the CTCs enumeration trials using flow cytometry technologies have made possible to implement these assays in certified clinical practice (CellSearch® system) [11]. The core techniques for the isolation and detection of CTCs are divided into two categories, immunological and morphological. Immunological techniques are based on the (CD45-), Epcam (+), panCK (+) epithelial origin phenotype of CTCs excluded (CD45+) white blood cells, with the main disadvantage of false negative results including losses from the epithelial-mesenchymal transition (EMT) and the Epcam (-), neoplastic cells; moreover false positive results may occur from macrophages epithelial antigen phagocytosis [12]. Morphological techniques are phenotype independent and based on the large size of CTCs in comparison with most blood cells, using specialized substrates perforated with micropores small enough to hold back the CTCs, while letting the blood cells through. Main disadvantage of morphological techniques is related to precision, repeatability and reproducibility quality control aspects necessary for the clinical diagnostics certification procedures [13].
In the present study, we designed and performed a combined methodology of isolation and detection of CTCs based on multiparameter flow cytometry and filtration cellular technology (ScreenCell® Technology) with clinical value in the patient's early diagnosis, prognosis and metastatic risk evaluation therapeutic procedures. For this purpose, we have established for the first time according to our knowledge, a novel protocol for the isolation of CTCs from peripheral blood based on a centrifugation and hydroxyethyl starch supplement sedimentation protocol using CD45 negative immunomagnetic separation system. In conclusion, this combined methodology could be a fast, simple and reproducible combined diagnostic procedure for the patients' personalized cancer treatment monitoring, via CTCs characterization and further DNA/RNA molecular analyses and ex vivo human CTCs functional assessment cultures.
A volume of 7.5 ml of peripheral blood (PB) from each patient (number of patients, N = 9) with different primary cancer type stage III metastatic disease were collected by trained phlebotomists of Oncology Clinic at Interbalkanean Medical Center of Thessaloniki with the first 2 ml being discarded to avoid possible contamination with skin epithelial cells. All patients were in good general condition and collection of peripheral blood has been processed before the programmed chemotherapy. Patient 1, male 75 years, diagnosed with a low differentiation adenocarcinoma oesophagus primary cancer (OC) with lymph nodes metastases, patient 2, female 51 years, a stage III Her2/Neu + breast primary cancer with sternum metastases (BC), patient 3, male 68 years, a stage III colorectal primary cancer (CRC) with lung metastases, patient 4, female 49 years, a stage III triple negative breast carcinoma (TNBC) and patient 5, male 70 years, a stage III invasive perivesical urothelial carcinoma (UC). Patients 6 to 9 have not been included in CTCs examination due to cut-off below counts of 1 CTC per 3 ml and considered negative for further analysis. An informed consent document was signed from all the donors according to Medical Doctor's guidelines. The collection tubes (BD® Vacuntainer K2E (EDTA) CE, 10.0 ml) were placed in specialized room temperature box. After collection and labelling of the tubes with consecu¬tive numbers and date and time of collection, the samples were stored at room temperature and transported to the clinical laboratory for immediate processing within 6 hours time from the collection time.
Fresh PB units (N = 9) centrifuged at x45 g RCF, 7 minutes, room temperature, brake off in 2% Hydroxyethyl starch supplement (Plasmin® Halexistar) final concentration following a second step of x500 g RCF, 15 minutes, room temperature, step to obtain a clear Total Nucleated Cells (TNC) fraction ("buffy-coat") as previous described [14]. The TNC fraction (1 ml from each PB unit) transferred aseptically with a Pasteur Pipette in a sterile 15 ml falcon tube (CellStar Tubes, Greiner bio-one) and diluted 1:1 by sterile Dulbecco΄s Phosphate Buffered Saline (1X) (Gibco by Life Technologies) in a 2 ml final volume. One million (1 × 10.6) of previous Red Blood Cells (RBCs)-depleted TNCs processed with anti-CD45 immumomagnetic beads (Mojosort® Human CD45 Nanobeads, Biolegend®) according to manual guidelines for the selection of (CD45-) negative fraction, prepared accordingly by following flow cytometry intracellular and cell surface staining protocol (Patent is pending).
The ScreenCell® filtration devices were developed in order to isolate CTCs by size on a microporous membrane filter. These devices are 19 cm long and designed for isolation of: i) Fixed cells for cytological studies (ScreenCell® Cyto); ii) Live cells for culture (ScreenCell® CC) and iii) Molecular biology (ScreenCell® MB). The PB was collected as described before and filtrated within three hours of collection. A volume of 3 ml PB was mixed with a dilution buffer for fixed cells and transferred into the ScreenCell® Cyto device for filtration.
CTCs multiparameter flow cytometric analysis: CTCs were analyzed for CD45, CD326 (EPCAM) and panCK expression percentage in total cells in FC500 flow cytometer (Cytometer FC500, Beckman Coulter, France) and for this purpose stained with anti-panCK-PE, anti-CD326-FITC and anti-CD45 PE/Cy5 according to cell surface and intracellular flow cytometry staining protocol quidelines (Biogenend®). Events acquisition was manually calculated on 150.000 events except patient 5 that calculated on 22.000 events due to sample loss during isolation procedure. Gating strategy was performed as to detect and quantify the CTCs as CD45-EPCAM + PANCYTO + immunophenotype cells percentage calculated on 7.5 ml total volume. For all patients post immunomagnetic isolation CD45-ve cells were calculated on 1.5 million cells per millilitre except patient 5 that calculated on 220.000 cells per millilitre. A volume of 100 microlitres per sample per patient stained with previous described monoclonal antibodies according the cell surface and intracellular staining manual of production company (Biogenend®). Data analysis was performed using, CXP 2.2 software (Cytometer FC500, Beckman Coulter, France). Aliquots of cells from patient 1 were also stained with isotype control antibodies.
CTCs microscopy analysis: CTCs isolated on the filter of the device were stained with May-Grunwald-Giemsa (MGG) in x40 microscope field of analysis. The filters were then mounted on a glass slide with Faramount mounting Medium (S3025, Dakocytomation, Glostrup, Denmark) and a circular glass coverslip. Under microscopy CTCs have nuclear size greater than or equal to 20 μm, a high and clear nucleocytoplasmic ratio (≥ 0.75), a dense, hyperchromatic nucleus, but not totally opaque and irregular nuclear membrane. Moreover, CTCs showing peripheral cytoplasmic vacuolisation with perinuclear clear space due to a minimal cytoplasmic retraction. Single CTCs may be encircled or infiltrated by platelets (tumoral microembol), by apoptotic cells during chemotherapy, by circulating giant macrophages with differential nuclei morphology and by cluster or solitary endothelial cells with nucleocytoplasmic ratio lower than that of CTCs leading to difficulties in differential diagnosis using CTCs. Cells enumeration was performed using a NIKON eclipse 50i fluorescence microscope integrated with CCD camera system and imaging software (NIKON, France) [15].
CTCs statistical analysis: CTCs flow cytometric and microscopy values are expressed and compared as mean ± standard deviation (SD) of six independent experiments from six donors. Statistical significance was determined by unpaired t-test statistical assay. The data derived are comparable to that obtained after referring to larger blood volumes (7.5 ml). CTCs value comparisons between the two analytic methodologies, the standard size-filtration and our multiparameter flow cytometry protocol, are considered not statistically significant according to related p values. CTCs values are considered statistically significant at p values < 0.05.
In order to obtain reliable results from our protocol, we firstly isolated CTCs from the same blood sample (3 ml), using the standard size filtration methodology already applied in clinical practice. Each PB sample (N = 9) was analyzed for the detection of CTCs microscopically using the ScreenCell® Cyto device with a cut-off of 1 CTC per 3 ml with 4 PB samples had a microscopic detection of 0 CTC per 3 ml and therefore not included in further multiparameter flow cytometric analysis. All patients aged 49 to 75 with a median of 62.6 years were histopathologically diagnosed with stage III metastatic disease as previously described and had performed a CTC detection prior receiving chemotherapy. CTCs that isolated and microscopically detected by ScreenCell® device ranged from 1 to 2 CTCs per 1 ml (3 to 6 CTCs per 3 ml) with a median of 1.60 and standard error ± 0.22 (Table 1, Figure 1) which is calculated from 7.5 to 15 CTCs per 7.5 ml with a median of 12.00 and a standard error ± 1.64 (Table 1, Figure 1). Notably, in Patient 3 (CRC) is detected a CTCs cluster under the microscopic evaluation of the ScreenCell® devices (Figure 2) and CTCs that isolated and previously microscopically detected followed by multiparameter flow cytometric analysis were ranged from 9.68 to 14.01 CTCs per 7.5 ml with a median of 12.10 with a standard error ± 0.78 (Table 2, Figure 3). CTCs per 1 ml and 7.5 ml value comparisons, between the two analytic methodologies, were considered to be not statistically significant since p-value = 0.9612, (Table 3). Finally, patients 6 to 9 have not been included in CTCs evaluation due to no detection of CTCs using Screen Cell® filtration device and considered negative for further flow cytometric analysis and CTCs value comparisons.
 Figure 1: Representative microscopic images A,B) of single CTC from patient 1 (OC); C) single CTC from patient 2 (BC); D) single CTC from patient 3 (CRC); E) single CTC from patient 4 (TNBC); and F) single CTC from patient 5 (UC) with well preserved morphology. MGG × obj.40 (see Materials and Methods).
View Figure 1
Figure 1: Representative microscopic images A,B) of single CTC from patient 1 (OC); C) single CTC from patient 2 (BC); D) single CTC from patient 3 (CRC); E) single CTC from patient 4 (TNBC); and F) single CTC from patient 5 (UC) with well preserved morphology. MGG × obj.40 (see Materials and Methods).
View Figure 1
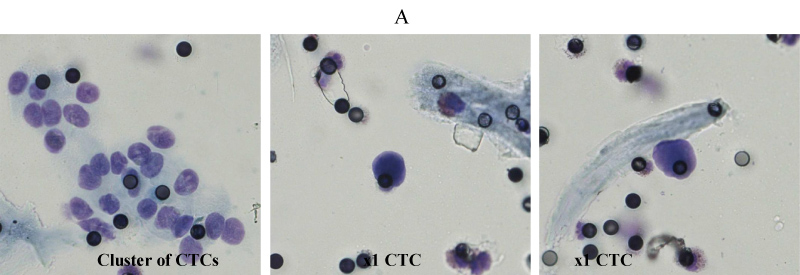 Figure 2: Representative microscopic images (A) of cluster of CTCs and 2 single CTCs from patient 3 (CRC).
View Figure 2
Figure 2: Representative microscopic images (A) of cluster of CTCs and 2 single CTCs from patient 3 (CRC).
View Figure 2
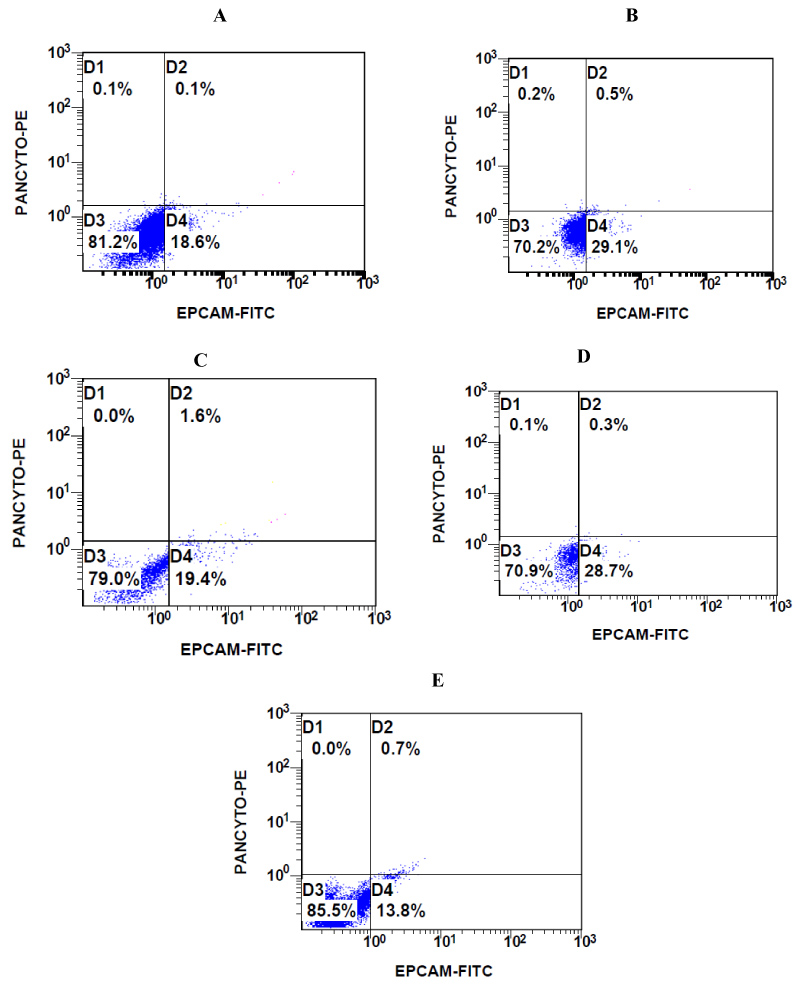 Figure 3: Representative flow cytometric images of CTCs from A) Patient 1 (OC); B) Patient 2 (BC); C) Patient 3 (CRC); D) Patient 4 (TNBC); and E) Patient 5 (UC) with CD45-EPCAM + PANCYTO + immunophenotype, gate D2 (see Materials and Methods).
View Figure 3
Figure 3: Representative flow cytometric images of CTCs from A) Patient 1 (OC); B) Patient 2 (BC); C) Patient 3 (CRC); D) Patient 4 (TNBC); and E) Patient 5 (UC) with CD45-EPCAM + PANCYTO + immunophenotype, gate D2 (see Materials and Methods).
View Figure 3
Table 1: CTCs microscopic quantification using size filtration ScreenCell® devices. View Table 1
Table 2: CTCs flow cytometric quantification using FC500 device. View Table 2
Table 3: CTCs microscopic (MIC) and flow cytometric (FC) quantification values comparison using unpaired two sample t-test, statistical significance p-value < 0.05. View Table 3
Tumor heterogeneity is observed in CTCs, which include subpopulations of differing aggressiveness and phenotypes [16]. The recent cellular technologies of isolation and characterization of CTCs are based on previous principles to reduce false positive and false negative results in trying to be harmonized according to Good Laboratory Practices (GLP) and Good Clinical Practices (GCP). Major current isolation technologies based on centrifugation, immunomagnetic and/or microfluidic selection of CTCs provides efficient post-processing cellular recovery in epithelial solid cancers with limitations on neoplastic cells that lose their differentiation epithelial antigens during epithelial-mesenchymal transition, due the possibility of phagocytosis of epithelial antigen by macrophages during tumor progression and in non-epithelial solid cancers. In contrast, Epcam-independent, based on morphology and size isolation techniques avoid previous problems with possible clinical applications also on non-epithelial cancers, such as metastatic melanoma, but with limitations on apoptotic and endothelial cells post-processing contamination due to CTCs size and morphology similarities.
By using liquid biopsies from blood, and through the identification of CTCs, the metastatic cells can be earlier detectable, before the clinical incidence of secondary tumors, a result that can permit the study of the molecular genomic and metabolic profile of these metastatic cells along with their chemosensitivity [17,18]. Moreover, considering CTCs monitoring and characterization technologies that are rapidly increasing in recent years, core molecular diagnostic and functional laboratory methodologies, such as microscopy, immunocytochemistry (ICC) and immunofluorescence (IF), 3-d fluorescence in-situ hybridization (3-d FISH), RT-PCR, digital droplet PCR, single cell next generation sequencing, mass spectrometry, multiparameter flow cytometry, 3-d functional tumor cell culture and tumorsphere assays and others, are aiming to upgrade and personalize therapy in the cancer healthcare sector [19-22]. In this manner, CTCs detection aspects, such as quality and quantity controls, cytopathological examinations, blood components infiltration, non-tumor and non-blood cells appearance, cellular malignancy criteria, clinical evaluation of uncertain malignancy and of cluster of malignant cells, tumor cell microenvironment analysis, needs to be investigated in the years to come, under carefully designed clinical trials.
Importantly, we were able to design a protocol and implement a novel combined methodology for isolation and detection of CTCs based on size filtration and flow cytometric technologies under advanced clinical laboratory standards, eliminating failures and limitations of related CTCs technologies. The results presented in this work have confirmed the accuracy of our methodology by applying a complementary approach that limits CTCs loss rates upon instrumentation, thus increasing the productivity and reliability of obtained data. Additionally, the combination of morphological and immunology technologies that used in our work giving a validated approach aiming to enhance the sensitivity of CTCs isolation and detections clinical outcomes. However, the previously described technologies have to further exploited in larger clinical trials in order to approve its clinical value and preciseness. In any case, however, the present study may be of high interest to clinical diagnostic laboratories and health professionals, such as oncologists, pharmacologists, biologists, geneticists, bioinformaticians and other specialists working on personalized (precision) cancer therapeutics field, aiming to increase significantly the therapeutic value of CTCs monitoring during tumor disease development and anticancer pharmacotherapy. Once the liquid biopsies and the CTCs isolation protocols permit the imaging and omics technologies to achieve their practical clinical utility, within the concept of precision cancer medicine, the routine CTCs screening for individual cancer patients will be globally achieved.
The authors would like to thank the Oncology Clinic at Interbalkanean Medical Center and especially Dr. Emmanouelides Christos for the generous assistance in the collection of CTCs. Moreover Screen Cell S.A and Biogenea pharmaceuticals Ltd. for kind scientific support. This research was carried out/funded in the context of the project "Molecular signatures analysis of three-dimensional cell cultures and circulating tumor cells in the treatment of cancer" (MIS 5004622) under the call for proposals "Supporting researchers with emphasis on new researchers" (EDULLL 34). The project is co-financed by Greece and the European Union (European Social Fund- ESF) by the Operational Programme Human Resources Development, Education and Lifelong Learning 2014-2020.
None.