Detecting the occurrence of pathogenic Leptospira spp. is vital for public health implications and epidemiological studies. Different isothermal-based techniques ascribed to loop-mediated isothermal amplification (LAMP) to detect Leptospira spp. in black rats and spiked environmental waters were evaluated. In this study, modified platforms were designed to detect the lipL32 gene encoding the major outer membrane lipoprotein of Leptospira spp. Standard and direct dry isothermal technique platforms exhibited high specificity. Analytical sensitivity was validated through 10.5 fg or 1 × 10-8 and 105 fg or 1 × 10-7, respectively. In spiked flood water, assays revealed favourable results. Overall, the dry LAMP technique developed has proven a confirmatory on-site test for the target Leptospira spp. present in its host, flood, and environmental waters suitably applicable in developing countries.
Isothermal amplification platforms, On-site biosensor, Leptospira spp, Environmental and floodwater
Leptospirosis goes by many other names, including Weil's disease, canicola fever, hemorrhagic jaundice, and mud fever. It is a zoonotic, rodent- and waterborne disease caused by pathogenic spirochetes of the Leptospira genus, a group of flexible, spirally shaped, obligately aerobic, Gram, negative bacteria. Leptospirosis is considered the most prevalent zoonosis in the world [1]. Humans frequently contract the infection via either direct or indirect contact with the urine of an infected animal. Warm climate countries are predisposed to have a significantly higher incidence than temperate regions [2] due mainly to the more prolonged survival of leptospires in the environment in warm, humid conditions. The disease has shown seasonal inclination, with peak prevalence occurring in summer or fall in cold-climate regions and during rainy seasons in warm-climate regions. In the Philippines, elevated cases of leptospirosis are reported from July to October every year. Within the country, the highest incidence is constantly found in the National Capital Region each year; this is attributable to several factors such as flood proneness, over-population, low socioeconomic status, and inadequate sanitation that favor exposure to sources of contamination and transmission the infection within the area. The usual entry point is through abrasions or cuts in the skin or the conjunctiva; infection can occur through intact skin after prolonged immersion in water. Waterborne transmission has been documented; point contamination of water supplies has resulted in several outbreaks of leptospirosis. Inhalation of water vapors or aerosols also may result in infection via the mucous membranes of the respiratory tract. Rarely, direct transmission between humans has been demonstrated; however, excretion of leptospires in human urine months after recovery has been recorded [3]. It is thought that the low pH of human urine limits the survival of leptospires after excretion. Transmission by sexual intercourse during convalescence has been reported [4,5].
On the other hand, wading in contaminated floodwater and ingestion of contaminated food or water are two typical modes of transmission in endemic areas. After entry into the host, pathogenic leptospires multiply in the renal tubules of chronically infected mammals [6]. This bacterium invades the human body through abraded skin and open wounds. It can also enter through the nostrils, mouth, or genitals and can be passed through intercourse or breastfeeding. Rodents and domestic mammals, such as cattle, pigs, and dogs, serve as significant reservoir hosts of the bacterium [7-7]. In addition, it can spread through the urine of dogs, rodents, and farm animals.
It is considered one of the most critical concerns in livestock, particularly cattle, due to reproductive failures such as abortion, embryonic death, stillbirths, weak offspring, decreased milk production, and growth rates [10-13]. The clinical course of leptospirosis is variable in humans. Some cases are mild, self-limited, or subclinical, while others are severe and potentially fatal. The illness generally presents with an abrupt onset of fever, rigors, myalgias, and headache in 75 to 100% of the patients, after an incubation period of 2 to 26 days (average 10 days). Leptospirosis has been described as a biphasic disease. The first phase consists of an acute febrile bacteremic phase lasting two to nine days, after which there may be a period with little, or no fever and apparent improvement. The second phase consists of an "immune" phase characterized by renewed fever and the development of complications that include jaundice and renal failure ("Weil's disease"), pulmonary hemorrhage, acute respiratory distress syndrome (ARDS), uveitis, optic neuritis, peripheral neuropathy, myocarditis, and rhabdomyolysis [14-16].
The Philippines is one of the Southeast Asian countries often affected by extreme typhoons and severe flooding. Due to the topographical attributes of the country - as it maintains around 421 river basins interspersed throughout the country and geographical location and - as it stretches across the Pacific Typhoon Belt, the Philippines has been predisposed to cyclonic storms in which frequency of occurrence and scale and intensity of intensity magnify each year. And adding salt to injury also wreaks a wide range of health consequences, the most common infectious and communicable disease that peak during higher rainfall than average and associated flooding is leptospirosis. Recent reports and reviews published are classified as one of the 17 Neglected Tropical Diseases (NTD) and reported as significant emerging and reemerging diseases by the World Health Organization. Locally, the disease inflicts an average of 680 cases and 40 deaths from the disease reported with a prevalence of 10/100,000 per year.
To date, several diagnostic methods have been described for the diagnosis of leptospirosis. These methods rely on the demonstration of the organism by microscopy and detection of antigens or nucleic acids in clinical samples such as blood, urine, milk or liver, kidney, and other tissue samples collected from maintenance and clinically affected hosts. MAT or microscopic agglutination test is considered the "gold standard" of serologic diagnosis. This assay requires ample time as it works by detecting the increase in antibody titer in serum samples obtained weeks apart. Although this technique provides an efficient retrospective diagnosis, it does not provide an early diagnosis [17]. Other diagnostic tests include dark-field microscopy, enzyme-linked immunosorbent assay, and Western blot and are known to have low sensitivity [18]. Polymerase chain reaction (PCR) and quantitative real-time PCR (qPCR) can be used to detect leptospirosis in clinical samples within the first week of the disease with low clinical sensitivity. Only about 50% of the culture and MAT confirmed cases were positive by qPCR [19]. Both PCR and qPCR are costly and often unavailable in many laboratories, especially those where leptospirosis is endemic.
Isothermal techniques such as LAMP or loop-mediated isothermal amplification are another type of nucleic acid-based detection system that has been progressively selected as a diagnostic tool of choice because of its wide array of applications and its flexibility of applicability even on less delicate test samples. It is a rapid, more specific, sensitive, and cost-effective method that can supersede many intricate molecular methods, including PCR. The capability of LAMP to detect point-of-care, pen-side, and even field-ready methods has been demonstrated in numerous works over the years. Therefore, this research aimed to corroborate the use of a loop-mediated isothermal amplification (LAMP) assay that detects leptospirosis in naturally infected rats and experimentally contaminated water sources. Specifically, this study attempted to devise various LAMP platforms that could become an accurate and precise field-based yet economical diagnostic tool whose essential steps towards a suitable LAMP assay like nucleic acid extraction and DNA amplification could be accomplished remotely from the laboratory where access to sophisticated equipment was not possible. Once proven effective, these assays would be recommended and adopted as potential tools for epidemiological surveys of leptospirosis in environmental and floodwater and for the surveillance and monitoring of the disease in high-risk populations in endemic and even non-endemic areas. For the first time in the country, the LAMP molecular technique demonstrated feasibility in leptospirosis-contaminated water sources. Therefore, it could be strongly suggested that LAMP could be utilized in acomprehensive field-based surveillance system for routine monitoring of Leptospira spp. in that way contributes to reducing human health risk for leptospirosis, especially in endemic and infected areas.
Thirteen (13) kidney samples were collected in the study to collect field-positive isolates of Leptospira spp. Before organ harvest, rats that were caged in households and rice fields were correctly sacrificed under the supervision of a licensed veterinarian. Kidney samples were collected aseptically from rats, placed in properly labelled microcentrifuge tubes, and kept at -200 °C until use for DNA extraction. According to the manufacturer's instructions, a standard commercial DNA elution method was used in all organs collected using the DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany). The purity and concentration of the resulting extracts were then determined using a calibrated spectrophotometer (IMPLEN Nano Photometer, Germany).
LAMP sets of primers were designed and generated based on the Leptospira spp. lipL32 gene using Primer Explorer (https://primer explorer.jp/e/) for LAMP optimizations, routine tests, and PCR detection using the outer primers F3 and B3 (Table 1) derived from the set of LAMP primers. The set of generated primers was based on the lipL23 gene of Leptospira interrogans serovar Manilae str. M933_lip32 NZ_AHPS01000269.1 encoding the major outer membrane lipoprotein, a virulence factor expressed in all pathogenic serovars of Leptospira.
Table 1: LAMP and PCR primer sets. View Table 1
PCR cycling condition was optimized in 10.0 μl reaction mixture prepared and mixed accordingly with the following reagents: 2 × Promega colourless buffer, 2 mM MgCl2, 1 mM dNTP mix, 1 uM each of F3 and B3 primers, and 5U of Promega GoTaq® polymerase enzyme. Amplification was carried outon a thermocycler (BIO-RAD, United States) with initial denaturation at 95 °C for 2 min followed by 35 cycles of denaturation at 94 °C for 30 min, primer annealing at 60 °C for 30 seconds, and DNA extension at 72 °C for 30 seconds before the final extension step at 72 °C for 5 minutes. The PCR products obtained after the assay were separated on 2.0% agarose gel (w/v), stained with SYBR Green I nucleic acid gel stain (Invitrogen, US). Electrophoresed using Mupid-One Electrophoresis System (Advance, Tokyo Japan) for at least 45 minutes set at 100 v and viewed with CLiNX Gel Documentation System (Clinx Science Instruments Co., Ltd, China). Samples that generated an approximate band size of 200 bp on the agarose gel after electrophoresis was noted for positive PCR reactions.
The amplification of leptospiral DNA was carried out using the generated sets of LAMP primers. The reaction was performed in a 12.5 μl reaction mixture prepared and mixed accordingly with the following reagents: 1.6 × Bst polymerase buffer, 1.2 M betaine, 0.2 μm and 0.8um each of outer (F3 and B3) and inner primer (FIP and BIP) respectively, 0.4 mM Intron dNTP mix, 6 units of Bst polymerase enzyme and 1.0 μl of DNA template. The standard LAMP mixtures were isothermally incubated in a mini dry bath incubator (HERCUVAN Lab Systems, United Kingdom) at 63 °C for 30 minutes. LAMP products were added with 1 μl of 1:10 (v/v) SYBR Green I (INVITROGEN) in each post-amplified tube and visualized under exposure to blue light. Products that emitted green fluorescence indicate positive LAMP reactions. Otherwise, orange to yellow was interpreted as negative. For confirmation purposes, LAMP products were also loaded in 2% agarose gel in which distinct ladder-like band patterns in positive LAMP products were noted.
For the dry LAMP platform, stabilization of all reagents was performed in a single step dry-up. All reagents of the optimized standard LAMP mentioned in the presence of a drying agent, trehalose, and a color indicator, the colorifluorometer indicator (CFI), were air-dried for 45 minutes on the interior walls of the PCR tubes. The LAMP mixture that was dried and stabilized in tubes contained 1.5 μl Bst polymerase buffer, 3 μl betaine, 1 ul of MgSO4 1.5 μl dNTP mix, 1 ul of primer mix (prepared by mixing 0.125 μl each of the outer primers F3 and B3 and 1.0 ul each of the inner primers FIP and BIP per reaction), 0.5 μl of the Bst polymerase enzyme, 1.5 μl of trehalose and 0.5 μl CFI. For the actual LAMP reaction, reconstitution and rehydration of the dried premixed LAMP were performed by adding 4 ul of DNA template and 8.5 ul of nuclease-free water that yielded a final assay volume of 12.5 μl. Isothermal incubation of rehydrated LAMP mixtures was done the same way as in standard LAMP. The sky-blue to light-blue color change is an interpreted positive result, while the violet color change was negative.
The analytical specificity of the developed techniques is defined as the unequivocal detection of an appropriate gene sequence of the target pathogens and differentiation from any other nucleotide sequence of non target pathogens or any other microorganisms present in the investigated samples. Standard and dry LAMP were tested and validated by molecularly challenging the purified sequence-confirmed nucleic acids of Leptospira spp. against the most common microorganisms in the environment that could also occur in rats, including Schistosoma japonicum, Staphylococcus spp., Salmonella spp., and E. coli.
In contrast, analytical sensitivity refers to the minimum number of copies in the sample that can be measured accurately, also called the limit of detection (LOD). To evaluate analytical sensitivity and establish LOD, sequence-validated nucleic acid from Leptospira spp. with known concentration was serially diluted up to 10 ten times in nuclease-free water.
A kidney sample harvested from a rat infected with leptospirosis was prepared for crude DNA extraction before actual spiking of DNA water using flood water. Two liters of floodwater were sampled from a garbage disposal area where rat sightings had been observed. The collected samples were divided into two equal volumes. One volume was kept as control, while the other was experimentally contaminated by the macerated kidney sample having a pre-identified quantity and concentration of DNA measuredby spectrophotometry. Subsequently, the samples were filtered using a Whatman Nucleopore membrane filter paper 1 (11 um pore size, 47 mm diameter, GE Healthcare Life sciences, UK). The water sediments retained on the paper were collected, transferred to 1.5 ml tubes with nuclease-free water, and centrifuged at full speed for 5 minutes to collect the pellet. As described, the resulting pellet was processed; DNA crudely extracted and assayed by PCR and LAMP. In the assay, triplicates of alkaline lysed macerated floodwater debris, uncontaminated floodwater, an off-target pathogen (Schistosoma japonicum), negative control with nuclease-free wateras a template, and positive control.
Figure 1 shows a representative PCR assay detection using the specific LAMP- F3 and B3 detection primers for the LipL32 gene of pathogenic Leptospira spp. With the optimized lipL32-PCR assay condition, 200-bp post-amplification target after gel electrophoresis. Ten out of 13 samples had positive reactions by PCR.
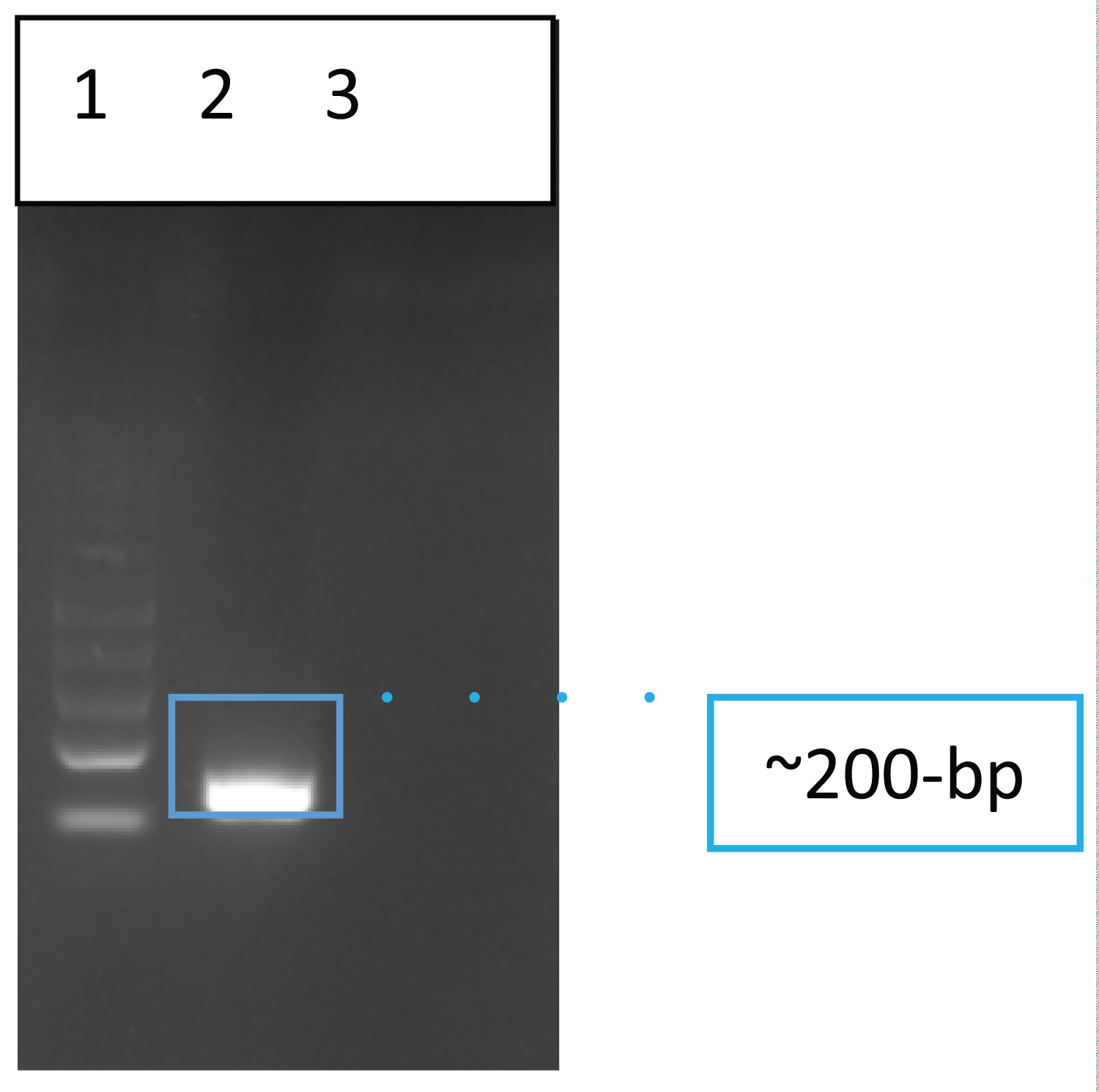 Figure 1: Gel electrophoretic result of the LipL32 PCR amplicon from a representative rat kidney extract. 100-bp Promega molecular ladder (lane 1), rat kidney sample (Lane 2), and Negative control- water as a template (Lane 3).
View Figure 1
Figure 1: Gel electrophoretic result of the LipL32 PCR amplicon from a representative rat kidney extract. 100-bp Promega molecular ladder (lane 1), rat kidney sample (Lane 2), and Negative control- water as a template (Lane 3).
View Figure 1
Standard and direct dry LAMP formats were successfully formulated for rapid molecular identification of Leptospira spirochetes in the rat kidney and flood and environmental water samples. Both formats were used in all 13 macerated kidney samples. Endpoint standard LAMP reactions resulting from incubation at 63 °C for 30 minutes generated positive amplifications observed with green fluorescence after incorporating a fluorescent dye and exposure to blue light and with the occurrence of distinctive ladder-like patterns in gel (Figure 2). As for direct dry LAMP, the actual drying method formed steady and well-attached purplish dried smudges at the bottom of the tube caps. The drying of the premixed LAMP reagents for 45 minutes also resulted in positive amplification (Figure 2). In addition, a post-amplification color change from purple to blue (sky blue to light blue, depending on the type of template used) corresponded with LAMP band patterns in the gel were also clearly demonstrated.
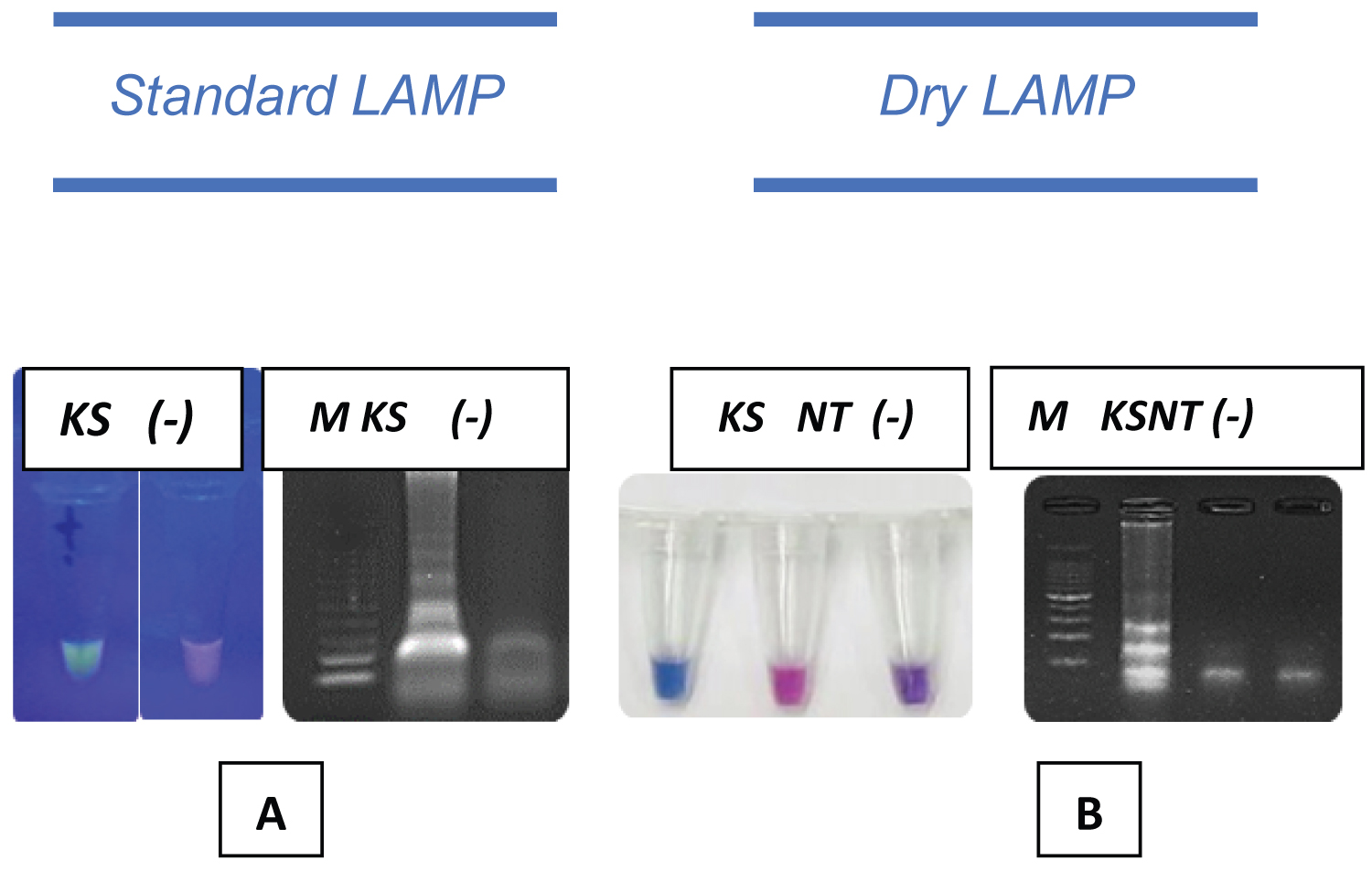 Figure 2: Standard and Dry LAMP Detection in a Rat Kidney Sample. Green fluorescence of the positive LAMP reaction after incubation at 63 °C for 30 minutes (A, Left). Gel electrophoretic analysis of LAMP products indicating positive results (A, Right). Dry LAMP results show blue color for positive reaction (B, Left); Purple to pink indicates negative reaction (B, Right).
Figure 2: Standard and Dry LAMP Detection in a Rat Kidney Sample. Green fluorescence of the positive LAMP reaction after incubation at 63 °C for 30 minutes (A, Left). Gel electrophoretic analysis of LAMP products indicating positive results (A, Right). Dry LAMP results show blue color for positive reaction (B, Left); Purple to pink indicates negative reaction (B, Right).
M: 100 bp Promega DNA molecular ladder; KS: Macerated Kidney Sample; NT: Non-Target DNA; (-): Negative Control
View Figure 2
The analytical specificity of oligonucleotide primer, lipL23, was tested against the four most encountered pathogens in rats and environmental and floodwaters. In addition, it was validated that molecular markers and techniques from recently described protocols were highly specific for Leptospira spp. All selected off-target pathogens included in the assay-pathogen challenge were tested negative against both LAMP platforms (Figure 3).
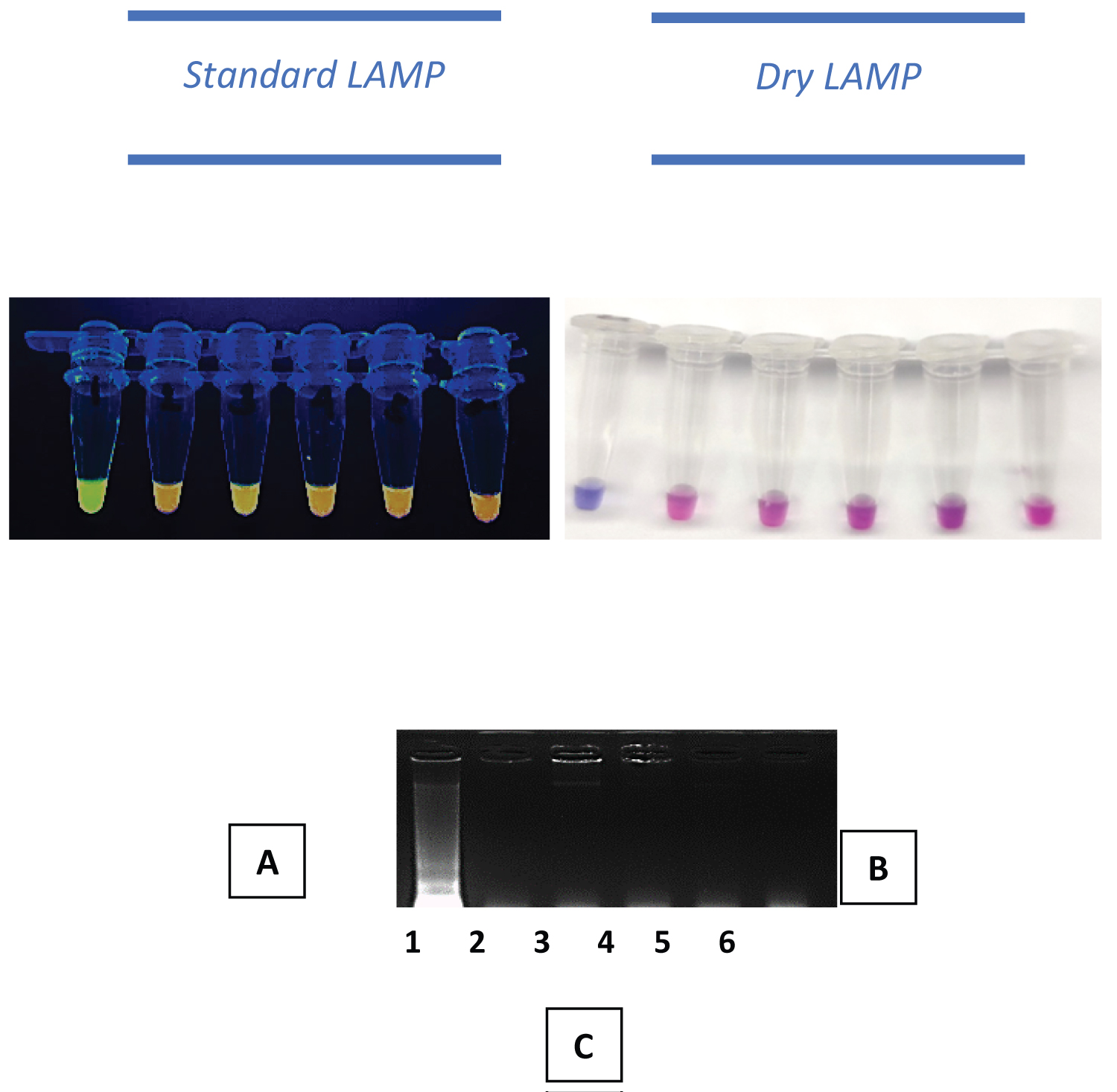 Figure 3: LAMP results of analytical specificity of standard and dry LAMP Assays. A: Standard LAMP by naked-eyed observation, green fluorescence indicates positive result (top); B: Results of dryLAMP by naked eye observation, blue reaction indicates positive, purple to pink, indicates negative reaction; C: Gel electrophoretic results showing ladder-like DNA bands from the positive reaction. 1) Leptospira spp. DNA 2) Schistosoma japonicum DNA 3) Staphylococcus spp. DNA 4) Salmonella spp. DNA 5) E. coli DNA and 6) Negative control.
View Figure 3
Figure 3: LAMP results of analytical specificity of standard and dry LAMP Assays. A: Standard LAMP by naked-eyed observation, green fluorescence indicates positive result (top); B: Results of dryLAMP by naked eye observation, blue reaction indicates positive, purple to pink, indicates negative reaction; C: Gel electrophoretic results showing ladder-like DNA bands from the positive reaction. 1) Leptospira spp. DNA 2) Schistosoma japonicum DNA 3) Staphylococcus spp. DNA 4) Salmonella spp. DNA 5) E. coli DNA and 6) Negative control.
View Figure 3
Assessments of the analytical sensitivity of previously described standard and dry LAMP protocols were performed by serially diluting a sequence-validated nucleic acid of Leptospira spp. in nuclease-free water. Initially, the concentration of undiluted macerated nucleic acid of Leptospira spp. was 1050 ng/ul, and after dilution, the concentration was as follows: 105 ng, 10.5 ng, 1050 pg, 105 pg, 10.5 pg, 1050 fg, 105 fg, and 10.5 fg and 1050 ag per ul. The results revealed that the detection limit of the LAMP platforms was detected down to 10.5 fg or dilution factor 10-8 for standard LAMP and 1050 fg or dilution factor 10-6 DNA per reaction for dry LAMP (Figure 4).
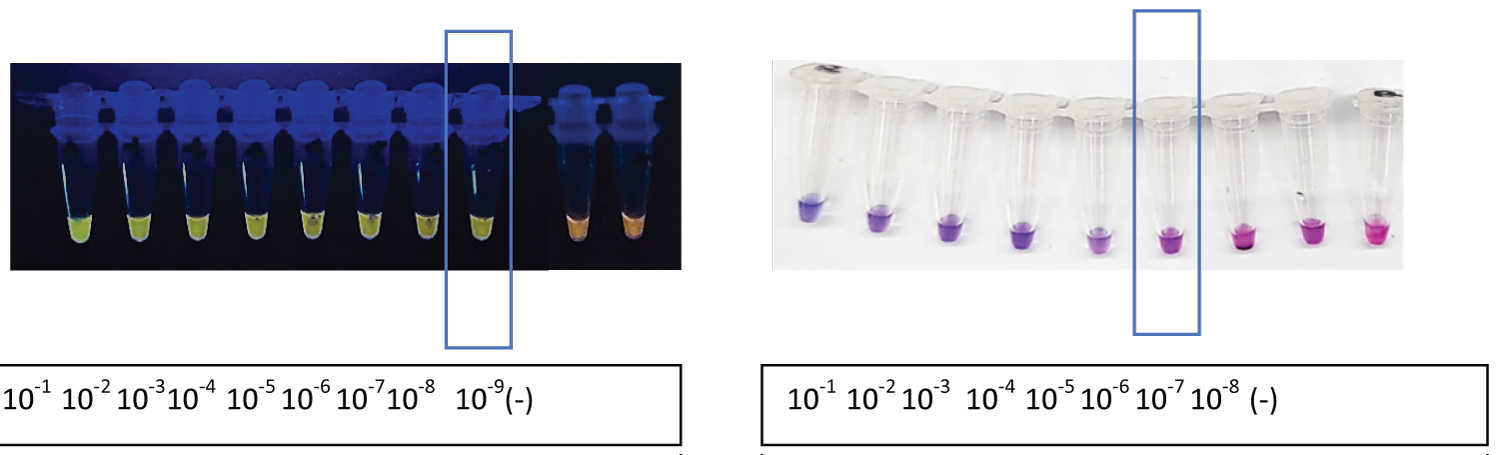 Figure 4: Analytical sensitivity of the standard and dry LAMP. The confined dilution in the boxed region indicated the point at which the least concentration of leptospiral DNA could be detected in an assay. Negative control (-).
View Figure 4
Figure 4: Analytical sensitivity of the standard and dry LAMP. The confined dilution in the boxed region indicated the point at which the least concentration of leptospiral DNA could be detected in an assay. Negative control (-).
View Figure 4
The detection rate related to the kidney samples assayed was compared by applying the PCR with detection primers (F3 and B3), the standard LAMP, and the modified dry LAMP assays. The detection rate was evaluated by computing the percentage rate involving the total number of positive samples for the assay and the total number of samples tested. Among the 13 samples tested, PCR had a detection rate of only 77% (10/13), while standard and dry LAMP assays both had 100%.
To represent an environmental condition where leptospires contaminate the water, developed PCR using the outer primers, standard and dry LAMP were tested using collected floodwater experimentally spiked with alkaline lysed kidney from a rat with naturally infected of leptospirosis. Results demonstrated that PCR, standard, and dry LAMP are comparable in detecting Leptospira species in the samples (Figure 5).
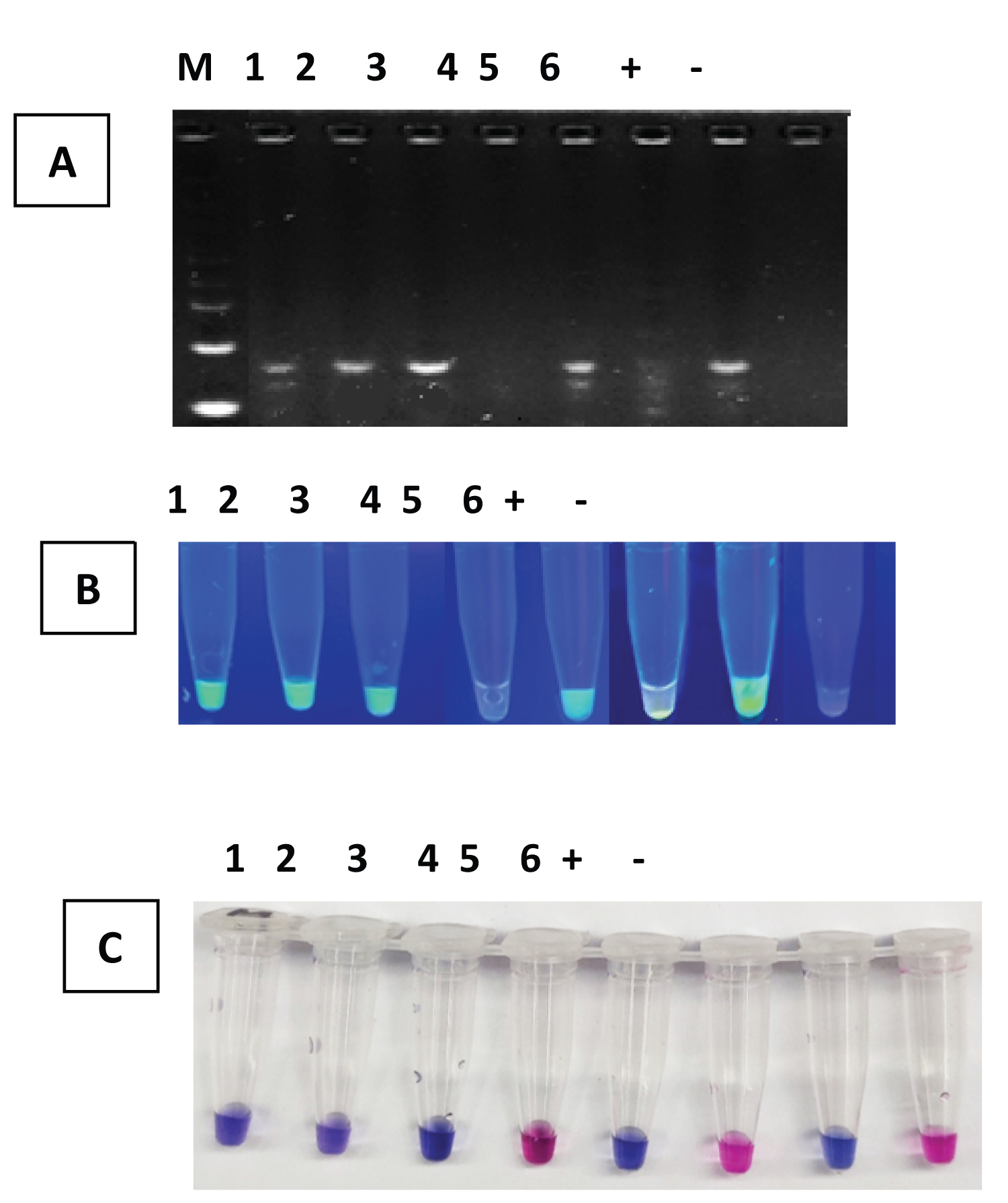 Figure 5: Different molecular platforms assay in leptospiral DNA spiked water sources. PCR, standard, and dry LAMP are comparable in detecting Leptospira species in the samples. A) PCR B) Standard LAMP and C) Dry LAMP. Leptospira-spiked water sources (1-6); Positive Control (+); Negative control (-).
View Figure 5
Figure 5: Different molecular platforms assay in leptospiral DNA spiked water sources. PCR, standard, and dry LAMP are comparable in detecting Leptospira species in the samples. A) PCR B) Standard LAMP and C) Dry LAMP. Leptospira-spiked water sources (1-6); Positive Control (+); Negative control (-).
View Figure 5
LAMP isothermal platforms were successfully evaluated in parallel with polymerase chain reaction to detect leptospirosis in rats and environmental and flood water. Standard and dry LAMP was effectively applied to analyze samples using crude extraction procedures without the complex DNA extraction procedures, implicating its potential in field-based applications. Developed assays enabled amplification of lipL32 gene, which encodes the outer membrane protein or subsurface lipoprotein that is only present in pathogenic strains of Leptospira spp., in collected kidney and spiked water samples. The detection rates for PCR, standard LAMP, and dry LAMP were 77%, 100%, and 100%, respectively, corresponding to the other studies suggesting that the LAMP platform is more sensitive than PCR.
One of the known drawbacks of LAMP is its dependence on the cold storage system during the assay, wherein in this study, another LAMP platform which is the Dry LAMP, cold storage of the LAMP reactions was excluded to produce stabilized and protected LAMP preparations while maintaining the efficacy of the operation of LAMP. Standard LAMP was modified as Dry LAMP by adding a protectant stabilizing LAMP reagents through a single-step drying procedure. In terms of the analytical performance of both LAMP assays, standard and dry LAMP, when tested against various off-target pathogens, showed high specificity for the target pathogen .Furthermore, analytical amplification sensitivities were validated to be up to 10.5 fg or 1 × 10-8 and 1050 fg or 1 × 10-6 DNA per reaction for standard and dry LAMP, respectively. Furthermore, both assays exhibited promising findings in simulated and spiked flood water.
In conclusion, the isothermal assay platforms were evaluated in parallel with PCR recommending the dry LAMP, which is specific, sensitive, stable, and, in general, faster, and also a reliable detection assay for pathogenic Leptospira species in clinical and environmental samples. The utility of dry LAMP targeting the lipL32 gene of Leptospira spp in this study has been initially applied in the Philippines. In general, the modified LAMP platform has proven itself to be a confirmatory test for the target pathogen present in its reservoir host and in floodwater that does not require sophisticated laboratory equipment, which is apotential molecular tool for the detection of Leptospira species in a field suitable for developing countries.
This work was supported by the Philippine Council for Health Research (PCHRD) and Development of the Department of Health (DOH) with a research grant: 416-306.