One of the main feature of Ebola virus is that Ribonucleic acid (RNA) plays more significant part instead Deoxyribonucleic acid (DNA) similarly as its hereditary material. When RNA is mirrored, many more mistakes are made than when DNA is mimicked [1]. This activates viruses like the Ebola viruses a predominantly high mutation rate when distinguished to DNA based viruses like smallpox or chickenpox. In the study of Ebola outbreak, experts found that the virus evolves much faster than it used to be in past. Scientists studying the epidemic found that the virus accumulates many mutations in the region of its genome that diagnostic tests use to recognize the disease. Too many mutations of this sort could render standard tests inaccurate, making it harder to diagnose Ebola infections and hampering the medical community's ability to treat and contain the virus. Changes such as these could also block vaccine development and therapies for treating the disease (Figure 1) [1].
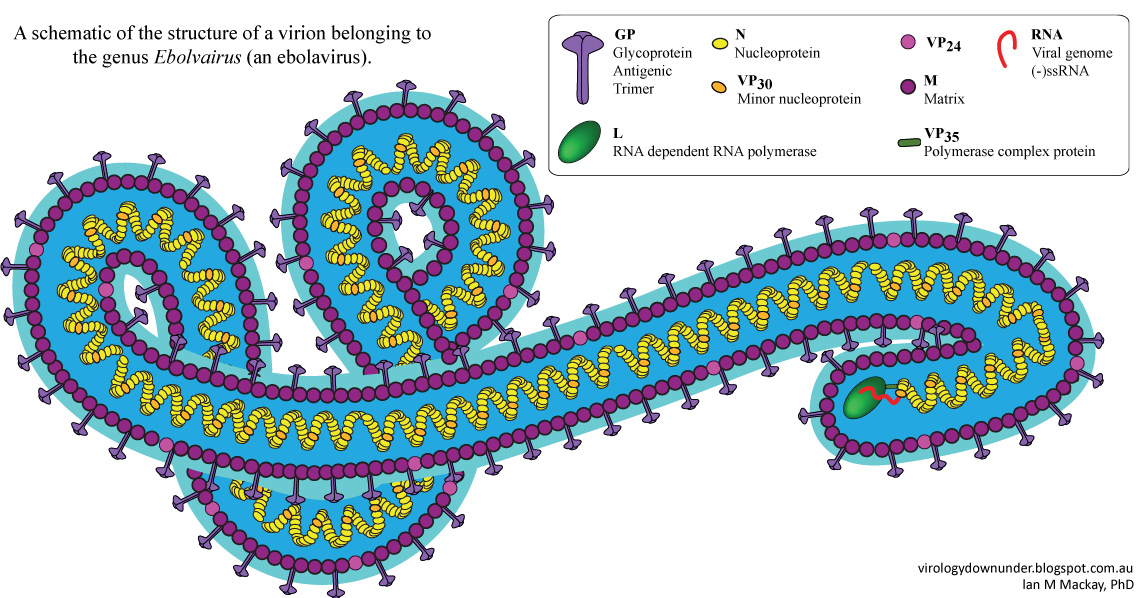 Figure 1: Schematic of the structure of a virion belonging to the genus Ebolavirus by Dr. Ian M Mackay [2].
View Figure 1
Figure 1: Schematic of the structure of a virion belonging to the genus Ebolavirus by Dr. Ian M Mackay [2].
View Figure 1
Scientists from National Institute of Allergy and Infectious Diseases (NIAID) compared EBOV-Makona isolates from initial outbreak; when certain mutations emerged in the viral surface glycoprotein and other unknown place. They infected mice and rhesus macaques with various mirrored virus that isolates to assess disease progression and viral shredding. Researchers also compared their investigation results from EBPV Makona isolates to findings from macaques infected with the original EBOV Mayinga strain from 1976. They found that EBOV (formerly designated Zaire ebolavirus) Makona is less virulent than EBOV Mayinga. The NIAID experts suggests intensifying studies on non-viral factors that may describe increased case numbers and fatalities. Cases include population mobility, available health care and virus tenacity in survivors – all may affect outbreaks dynamics.
"They were unable to find any vital divergence between early and late isolates lacking or carrying those mutations, suggesting that these mutations do not lead to variations in the disease causing ability in animal prototypes" Figure 2 [3,4].
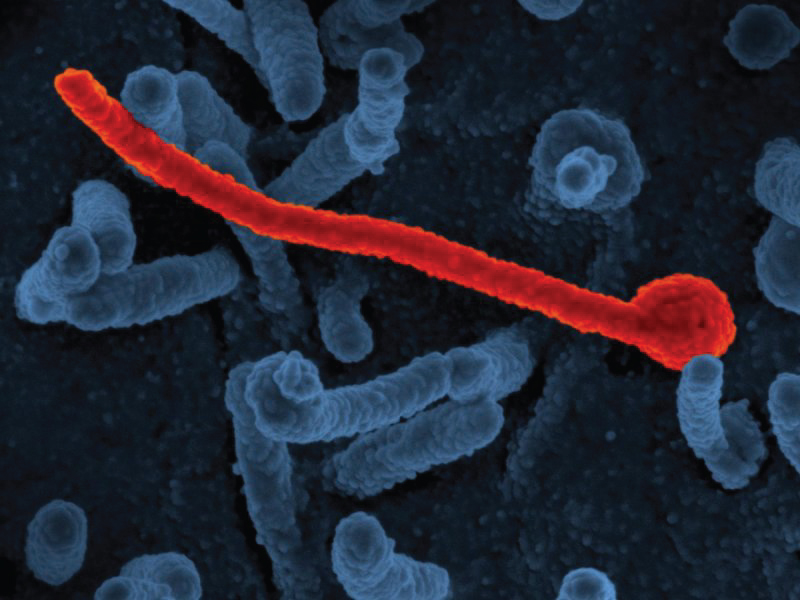 Figure 2: Electron micrograph of Ebola virus Makona (in red) from the West African epidemic shown on surface of Vero cells (blue) Image Credit: NIAID [4].
View Figure 2
Figure 2: Electron micrograph of Ebola virus Makona (in red) from the West African epidemic shown on surface of Vero cells (blue) Image Credit: NIAID [4].
View Figure 2
Marburg and Ebola viruses cause hemorrhagic fever in humans and other living organisms. About 50 to 90 percent of cases lead to death due to strains and infection, also there was no cure or alternate viable medication. Maury led a multidisciplinary with the team; they used a new bioinformatics based approach developed by John Chiorini at NIDCR, to recognize a protein called TIM-1 as a receptor for Ebola and Marburg viruses. Successive experiments demonstrated that both Marburg and Ebola viruses use TIM-1 as the receptor for infecting cells. The research also demonstrated that TIM-1 protein is widely expressed on epithelial cells that line different tissues in the body including mucosal surfaces of the airways and in the eyes. Maury also noted that these locations are regular with some of the ways the Ebola virus is thought to be spread through inhalation of aerosolized droplets and hand to eye contact.
The result demonstrated that being able to block Ebola's virion/viral particle entry into epithelial cells perhaps with a human matched version of the ARD5 antibody, may provide a way to stop initial infection and potentially limit the speed of the disease during an outbreak. And most importantly, study demonstrated that TIM-1 protein is not expressed on all the cell types that are infected by Marburg and Ebola.
"It's clear that there are receptors for Ebola because when TIM-1 was found on a number of epithelial cells in the body, it was not found in some important cell types that are contaminated by Ebola" was confirmed by Maury (Figure 3) [5,6].
 Figure 3: Illustrates Cellular protein TIM-1 acts as a receptor for Ebola virus and Marburg virus. Microscope image shows TIM-1 expression (in green) on the surface of human airway cells. Image Credit: University of Iowa [6].
View Figure 3
Figure 3: Illustrates Cellular protein TIM-1 acts as a receptor for Ebola virus and Marburg virus. Microscope image shows TIM-1 expression (in green) on the surface of human airway cells. Image Credit: University of Iowa [6].
View Figure 3
Ebola virus has seven genes, one of them is called GP, codes for two related proteins: a shorter secreted one and a longer one that extents the viral wall and sticks out of its surface. Throughout virus infection, some of the surface GP is cut off and is subsequently shed from contaminated cells. High levels of both shed and secreted GP are originated from the blood of infected humans and animals. Scientists [7,8] found that shed but not secreted GP that can bind to immune cells that are called macrophages and dendritic cells both are the targets of Ebola virus infection. Upon binding of shed GP, these immune cells start freeing massive quantities of immune modulators. Both shed GP and these immune modulators are soluble proteins that may transmit in the blood stream and this may demonstrate how continuous creation and release of shed GP an original immune response to the virus gets more worst and leading to high fever, massive inflammation and possibly the jolt that kills many Ebola patients.
Researchers [7,8] also found that the effects of shed GP on the immune cells depends on molecule called TLR-4. Blocking TLR-4 with specific antibodies former to exposing immune cells to shed GP damped the cell's reaction and eradicated much of the release of immune modulators.
Besides massive and destructive inflammation, lethal Ebola disease is also accompanies with loss of blood vessel integrity hence the name Ebola hemorrhagic fever (Figure 4) [7,8].
 Figure 4: Illustrates the role of shed GP during Ebola infection. Image Credit: Volchkov, et al. CC-BY [8].
View Figure 4
Figure 4: Illustrates the role of shed GP during Ebola infection. Image Credit: Volchkov, et al. CC-BY [8].
View Figure 4
When cells are infected with Ebola virus, the cell is reproduced by the virus and turned into a production factory that makes massive amounts on new virions. These virions are then removed from the cell to invade other cells and promote spreading infection. Tetherin is one of the immune system's responses to viral infection. If it works properly, tetherin prevents the contaminated cells. Conversely, the study [9] shows that the Ebola virus has advanced a new way to disable tetherin, thus blocking the body's immune system and allowing the virus to spread other cells. Past studies found that tetherin plays a very vital role in immune system's response to HIV-1, a retrovirus, and that tetherin is also obstructed by HIV. These new studies [9] disclose that human cells also use this defense against other types of viruses such as Ebola that are not even closely related to HIV-1.
"This information provides new way to study how tetherin works. Binding of a protein created by Ebola to tetherin clearly inactivates this contaminated cellular factory. Understanding how the Ebola protein prevents the action of tetherin, allowing the cells natural defense systems to slow down viral replication and provides the person or animal a chance to mount an effective antiviral response and recover" Figure 5 [9].
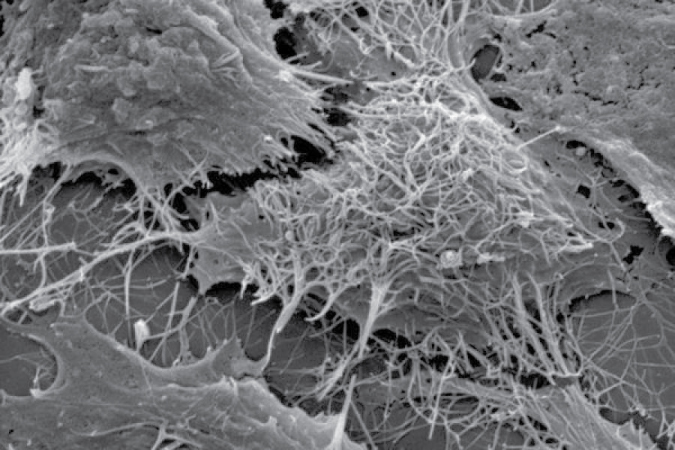 Figure 5: Illustrates scanning electron microscope image of Ebola virions (spaghetti-like filaments) on the surface of a tetherin-expressing cell (center). Image Credit: Paul Bates, PhD, University of Pennsylvania School of Medicine [9].
View Figure 5
Figure 5: Illustrates scanning electron microscope image of Ebola virions (spaghetti-like filaments) on the surface of a tetherin-expressing cell (center). Image Credit: Paul Bates, PhD, University of Pennsylvania School of Medicine [9].
View Figure 5
Ebola virus integrates a cellular enzyme into its virus particles, simplifying the infection to the target cells. When enzyme Xkr8 is activate, it flips a phospholipid called phosphatidylserine (PS) from the inner layer of the Ebola virus membrane (envelope) to the other layer. The wide-open PS enables entry of the virus. Scientist at Hokkaido University and the University of Tokyo generated ebolavirus like elements by expressing viral proteins in cultured mammalian cells to examine mechanisms by which Ebola virus enters target cells. When scientist blocked the activation of enzyme Xkr8, the exposure of PS on the surface of the virus elements was reduced. In Ebola virus contaminated cells, the virus components replicate and accumulate to produce progeny viruses. The progeny viruses bud from the surface of host cells, acquires an envelope derived from the host's cell surface membrane. Scientist have identified a key procedure that enables the Ebola virus to contaminate host cells, providing a novel target for developing antiviral drugs (Figure 6) [10-12].
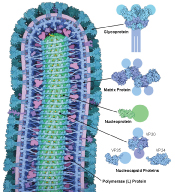 Figure 6: Illustrates Cross section through Ebola virus demonstrates proteins in blue, green and magenta, the RNA genome in yellow, and the membrane in light purple. Atomic structures are shown on the right, with portions that have not been determined shown with schematic circles [12].
View Figure 6
Figure 6: Illustrates Cross section through Ebola virus demonstrates proteins in blue, green and magenta, the RNA genome in yellow, and the membrane in light purple. Atomic structures are shown on the right, with portions that have not been determined shown with schematic circles [12].
View Figure 6
Author would like to thank Prof. Navarun Gupta, Prof. Hassan Bajwa, Prof. Linfeng Zhang and Prof. Hmurcik for their academic support. Author also thanks anonymous reviewers for their comments.
There are no conflict of interest as per Author's point of view.