As of 18 March 2020, more than 191127 cases of confirmed COVID-19 have been documented globally with over 7807 deaths. The impact of COVID-19 on patients with HIV have common presentations like non-HIV patients include fever, dry cough, difficulty of breath. Parallel evidence indicates that the risk of disease increases with critically more than 66 years age, including chronic lung disease. It is very difficult to use drugs on the market. We apply drug design computer techniques to find the expected drug for COVID-19 after knowing the repurposing the detailed of 3D-structures of its key proteins. Several known drugs act as strong inhibitors of COVID-19 protease, including: Abacavir, Lamivudine and Zidovudine. The combination of the analyzed spectra of the antiviral drugs: Abacavir, Lamivudine and Zidovudine help to give an additional information about the investigated set of complex drugs.
Coronavirus, COVID-19, HIV drug combination, Trizivir
As of 18 March 2020, more than 191127 cases of confirmed COVID-19 have been documented globally with over 7807 deaths [1].
we reported previously a computational modeling drug design for a category of HIV drugs called non-nucleoside reverse transcriptase inhibitor (NNRTI), nucleoside reverse transcriptase inhibitors (NRTIs) and protease inhibitors (PIs), respectively [2,3].
Following these publications, we published other articles describe the antitumor activity of a series of photosensitizers tested for several cancer cell lines, and in vivo experiments. We received positive feedback about our research [4-6].
Patients with COVID-19 and HIV are more susceptible to infection due to their Immunodeficiency status. For this reason, all researchers who interest with health care should understand the situation of the disease and find the selective drug to avoid the spread of the disease.
In this work, we applied multiscale modified non-invasive computational modeling techniques to discover drugs that may be used for repurposing the target COVID-19 protease.
We use drug combinations of three neutral models of Nucleoside/Nucleotide Reverse Transcriptase Inhibitors (NRTIs) Anti-Viral drugs: Abacavir, Lamivudine and Zidovudine (sold under the brand name Trizivir) [7] in comparison with another drug combination as described in Table 1 [8,9].
Table 1: In vitro/in vivo efficacy of the HIV drugs selected for COVID-19. View Table 1
Trizivir is used widely for controlling Human Acquired Immunodeficiency Syndrome. In order to extract the reliable information. We subject these data to the procedure of the optimal linear smoothing (POLS), in addition to 3-dimensional Multidimensional Scaling (MDS) methods [3]. Due to the importance of these three drugs for the medical treatment, we compare the cosine correlation through a 3 × 3 matrix of comparison of all drugs provide visualized information of the three drugs. Figure 1 2D-Structures of neutral expected antivirals repurposed drugs.
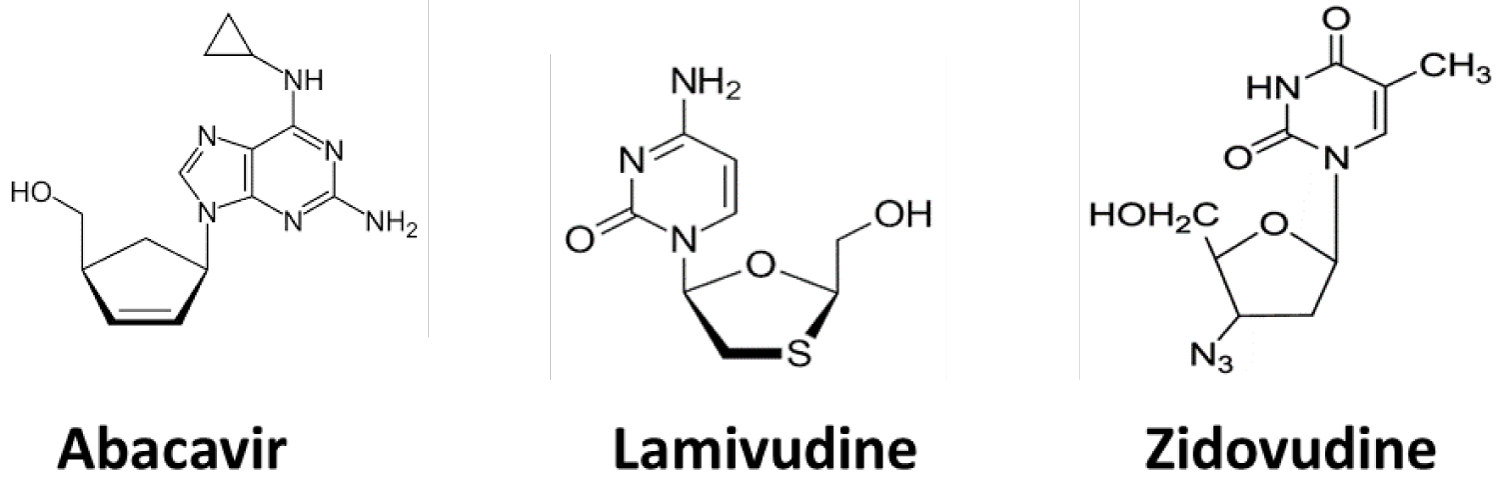 Figure 1: 2D-Structures of neutral expected antivirals repurposed drugs. View Figure 1
Figure 1: 2D-Structures of neutral expected antivirals repurposed drugs. View Figure 1
Combinations of Abacavir, Lamivudine and Zidovudine with these direct-acting antivirals could reduce viral replication, and the aberrant host inflammatory response and viral infectivity.
This work demonstrates that the use of an Al-driven analyzed data can facilitate rapid drug development, compared to the experimental methods, computer-modelling drug approaches are efficient in providing possible drugs for epidemic disease like COVID-19.
In this study, we use the crystal structure of COVID-19 protease. Abacavir, Lamivudine and Zidovudine used as model for neutral antiviral drugs, may identified to have inhibitory activities against COVID-19 protease. Our computational study can facilitate us to design novel inhibitors targeting COVID-19.
The authors declare that there is no conflict of interest.
There was no funding for this research.