International Journal of Brain Disorders and Treatment
Maternal Undernutrition in Rats Affects Hippocampal Development through Inhibition of Neurogenin 1
Marquis L Jessie1, Lili C Kudo2, Nancy Vi2, Kimbley Lau3, Mina Desai1, Michael G Ross1* and Stanislav L Karsten2,3
1Department of Obstetrics and Gynecology, Los Angeles Biomedical Research Institute at Harbor UCLA Medical Center, USA
2Neuroindx, 1655 E 28th Street, Signal Hill, CA 90755, United States
3Department of Neurology, Division of Neuroscience, Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center, USA
*Corresponding author:
Michael G Ross, MD, MPH, Department of Obstetrics and Gynecology, Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center, 1000 W Carson St., RB3, Box 467, Torrance, CA 90502-2006, USA, E-mail: mikeross@ucla.edu
Int J Brain Disord Treat, IJBDT-2-013, (Volume 2, Issue 2), Research Article; ISSN: 2469-5866
Received: February 19, 2016 | Accepted: July 08, 2016 | Published: July 11, 2016
Citation: Jessie ML, Kudo LC, Vi N, Lau K, Desai M, et al. (2016) Maternal Undernutrition in Rats Affects Hippocampal Development through Inhibition of Neurogenin 1. Int J Brain Disord Treat 2:013. 10.23937/2469-5866/1510013
Copyright: © 2016 Jessie ML, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Maternal undernutrition (UN) in rats inhibits neurologic development in offspring. To investigate these alterations we compared hippocampal global gene expression profiles of UN and control newborn pups. Whole transcriptome microarray analysis revealed significant changes in the expression of several genes including Neurogenin1 (Ngn1), a known neuro developmental regulatory factor. Expression of Ngn1 in the UN hippocampus was nearly 70% reduced. Western blot analysis using antibodies against Ngn1 protein, also showed more than a 50% decrease in the protein expression of Ngn1 in the hippocampus of UN newborn pups. Previously it was demonstrated that Ngn1 expression may be controlled through Notch pathway feedback inhibition. Accordingly, we found that Notch 1 protein expression demonstrates significant increase in UN newborn hippocampus compared to controls (CNTR 95.7 ± 28.0, UN 198.8 ± 66.8, P < 0.05). Analysis of neuronal cell counts using NeuN immunohistochemistry did not show a decrease in newborn pyramidal cells in UN pup hippocampus compared to controls. However, when NeuN immunohistochemistry was performed on postnatal day 21 (P21) pups, there was a statistically significant decrease in pyramidal cell numbers that was specific to the CA2/3 region (CNTR 180 ± 14, UN129 ± 3, P < 0.05). These data indicate that maternal UN causes a global reduction in offspring Ngn1 production and expression which may be linked to Notch1 inhibition. This, in turn, may lead to decreased neuronal number or altered regional distribution of hippocampal neurons. We propose that a predisposition to adult hippocampal disease may result from developmental effects of the in utero environment.
Keywords
Ngn1, Hippocampus, Food restriction, Growth restriction, Programming
Introduction
Maternal undernutrition in humans may result in intrauterine growth restricted (IUGR) offspring, which exhibit a predisposition toward adulthood disease [1]. This concept, known as intrauterine programming, stems from the work of David Barker (the “Barker Hypothesis”) who described an increased incidence of cardiovascular disease in association with low birth weight in humans [2].
Epidemiologic studies have shown links between IUGR and adulthood obesity, diabetes, cancer and neurodegenerative diseases such as Alzheimer’s disease [3-8]. Animal studies in multiple species have confirmed the association of IUGR (often produced by maternal under nutrition; UN) with low birth weight offspring which develop adult obesity [9] and metabolic syndrome. Furthermore, low birth weight offspring also display an array of neurologic, cognitive and behavioral abnormalities including increased food intake [10]. These findings raise the question as to whether there is a common fetal factor associated with both obesity and neurologic disease.
Studies from our laboratory have confirmed that maternal UN impairs fetal neuro progenitor cell migration, proliferation and differentiation [3] in association with altered hypothalamic development [4]. In view of these results, it is possible that maternal UN-induced low birth weight affects offspring brain development in a way that may predispose towards adult neuropathology.
To assess the neuro-molecular factors induced by maternal UN our present work utilized microarray based approach to examine the global transcriptome of the developing hippocampus. We found several genes that were either up or down-regulated in low birth weight pups. Neurogenin1 (Ngn1) was among those genes which were down-regulated. Ngn1 plays a central role in neuronal differentiation. It is a member of the neurogenins which are a family of basic helix-loop helix (bHLH) transcription factors essential in sensory neuronal development. Ngn1 specifically acts as a neuronal differentiation regulator, binding and enhancing the expression of genes that encode transcriptional regulation of neurogenesis. Our results demonstrated alterations in Ngn1 and its upstream regulator Notch1, which may represent the mechanistic pathway underlying evidence of reduced hippocampal neurons.
Materials and Methods
Intrauterine growth restricted rat model and tissue processing
Our maternal UN model was used as previously described [5]. Briefly, first-time pregnant Sprague Dawley Rats (Charles River Laboratories, Hollister, CA) were housed in a facility with constant temperature and 12 hour light to dark cycle. Rats were fed either a standard diet of normal laboratory chow or a 50% reduced quantity diet of the same laboratory chow. This diet was initiated from pregnancy day 10 and continued until delivery. Both control and UN dams were fed congruent diets of normal laboratory chow before this time period. The rat pups were allowed to deliver normally. On the day after delivery, litters were limited to four males and four females, which were nursed by ad libitum fed dams. Pups in excess of eight per litter were sacrificed on postnatal day 1 (P1).
P1 male offspring were sacrificed by rapid decapitation and whole brains were removed. The hippocampi were then dissected from twelve P1 pups (six UN and six control; one pup per litter) and stored at -80° for mRNA and Western blot studies. Total RNA was extracted using acid phenol extraction and the concentration and quality of RNA was determined using Nanodrop spectrophotometer and confirmed on Agilent bioanalyzer. For immunohistochemistry studies, additional animals were studied as follows: whole P1 (3 UN and 3 control) and P21 (4 UN and 4 control) brains were dissected, separated from the spinal cord after decapitation and quickly frozen at -80°C.
RNA isolation, amplification and labeling
For microarray gene expression experiments, four UN and four control (one pup per litter) was male P1 pups were used. Total RNA was extracted using acid phenol extraction (Trizol LS; GIBCO/BRL). The concentration and quality of RNA were determined using the Nanodrop spectrophotometer and confirmed on the Agilent Bioanalyzer. Agilent Whole Rat Genome microarrays were used for comparisons between UN and normal control hippocampal transcriptomes. Five hundred nanograms of total RNA were used for one round T7-based amplification and labeling protocol (Agilent Linear Amplification Kit PLUS). Dye incorporation efficiency was checked with an ND-1000 spectrophotometer (NanoDrop Technologies, Rockland, DE) and the Agilent Bioanalyzer 2100. Each microarray replicate was repeated twice with dye flip to correct for unequal dye incorporation rates. Therefore, eight microarray hybridizations were performed in total. Raw microarray data were acquired using the Agilent DNA Microarray scanner and processed with the accompanying Agilent Feature Extraction 10.5 Image analysis software using default settings (see MIAME report). Global lowess normalized signal intensities were used to identify gene expression changes in IUGR hippocampus. For the identification of differential expression, the genes were required to pass two conservative criteria: a ratio beyond the 95% confidence interval observed in homotypic comparisons, which corresponded to an approximately 1.5-fold expression change, and a paired t-test (P < 0.01) [6-8]. (Please see statistical analysis under results for more details).
Western blot analysis
Western blots were performed as previously described5using six UN and six control (one pup per litter) male rats. Briefly, previously frozen P1 hippocampal tissue was weighed and sonicated in protease inhibitor cocktail with hypotonic buffer. Using supernatant from these sonicated samples and Pierce BSA albumin standard curve, protein concentrations were calculated and samples containing approximately equal amounts of protein concentrations were loaded. These proteins were separated by Tris-HCL gel and transferred to PVDF membranes. The membranes were incubated overnight in either primary anti-body with blocking buffer of 1) rabbit anti-rat Ngn1 (1:1000 Goat anti-rat Santa Cruz Biotechnology, Inc) or 2) mouse anti-rat Notch1 (1:250 Mouse anti-rat Invitrogen Corporation). In all cases there was equal protein. They were then conjugated to the appropriate secondary antibody (either goat anti-rabbit or anti-mouse Bio Rad Laboratories, Inc). Signal was captured using chemiluminescence (Amersham ECL Kit). Immunoblots were imaged by scanning autoradiographs using Umax GS800 Calibrated Densitometer and Quantity One program. Resulting raw pixel density values were imported into Microsoft Excel for statistical analysis (Please see statistical analysis under results for more details).
Immunohistochemistry
Immunohistochemistry was performed as previously described [9]. Briefly, sagittal slices of P1 (3 CNTR and 3 UN) and P21 (4 CNTR and 4 UN) cortex containing hippocampus were mounted on Vectabond-coated glass slides and stored at -80°C. The slides were then prepared by first washing three times with 0.1 M TBS (with 0.5% triton, TBST). Each slide was then blocked for nonspecific binding with 10% donkey serum in TBST for 2 hours at room temperature. They were then incubated overnight with primary antibody for Neu N (1:250) with TBST at -4°C. The slides were again rinsed three times with TBST and incubated for 2 hours in the appropriate secondary antibody (1:125) with 10% donkey serum-TBST at room temperature. The slides were them washed three times with TBS and the signal was detected by Vector anti-fade medium. All sections were examined and digital images taken using Zeiss microscope (Axioskop) equipped with a Zeiss digital camera (Axiovision). Neurons from specific hippocampal regions were then manually counted for statistical analysis.
Statistical significance was determined using a two-tailed student’s test assuming unequal variance with significance determined by P < 0.05. (Please see statistical analysis under results for more details).
Results
UN hippocampal transcriptome
To test if UN alters the global transcriptome of the developing rat brain and identify potential regulatory factors involved, we used microarray based approach to investigate global hippocampal gene expression. Microarray data analysis revealed that a limited number of genes are specifically altered in the UN hippocampus with 18 known genes (Table 1) and 16 expressed sequence tags (ESTs). Only one gene, Ngn1, represented a known regulatory protein involved in hippocampal neuronal development. Ngn1 demonstrated a near 2-fold decrease on microarray analysis in the UN hippocampus compared to normal controls.
![]()
Table 1: Genes regulated in UN hippocampus. IUGR column shows average fold change between UN and normal controls. n - number of probes detecting the same genes in the experiment. Ngn1/Neurod3 (in yellow) shows a near 2 fold decrease (-1.7) in mRNA expression.
View Table 1
To confirm that this decreased expression of Ngn1 mRNA translated into decreased Ngn1 protein expression, we performed Western blot analysis of hippocampal tissue in both control and UN P1 rat pups. We observed a marked difference in Ngn1 protein production in UN pups compared to controls (Figure 1A). When these data were normalized to β-tubulin, similar to our microarray data, we observed a 50% decrease in Ngn1 protein production in UN pups compared to controls.
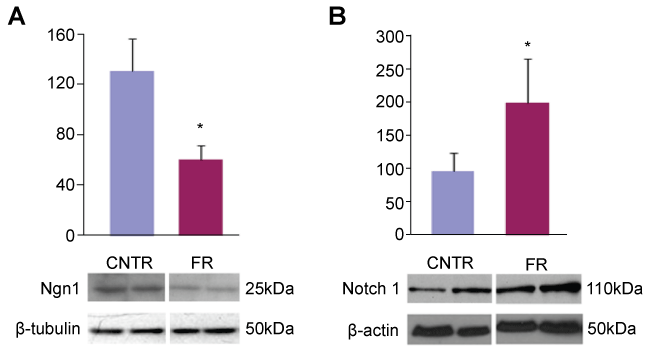
.
Figure 1: Ngn1 and Notch 1 expression in P1 rat hippocampus. A: Western blot of P1 rat hippocampal cell lysates probed for Ngn1. All data are mean ± SEM (n = 6) *P < 0.05; B: Western blot of P1 rat hippocampal cell lysates probed for Notch1. All data are mean ± SEM (n = 4) *P < 0.05.
View Figure 1
Also of note, tryptophan hydroxylase, an enzyme integral in serotonin (5HT) synthesis, similarly showed a greater than 2 fold (2.3) decrease on same microarray analysis when compared to controls (Table 1). Serotonin plays a key role in human affect disorders, including chronic depression [10], as noted in the discussion below.
Notch pathway is altered in UN rat hippocampus
Ngn1 is controlled by lateral inhibition through the Notch pathway [11]. To establish evidence of this control in UN pups, we performed Western blot analysis of hippocampal tissue from UN and control rats using Notch1 primary antibody. Figure 1B shows nearly a two-fold increased Notch1 production in UN pups compared to controls.
Neuronal cell distribution is altered by UN in P21 pups
Immunofluorescence targeting NeuN antigen were performed on hippocampal sections of both UN and CNTR P1 and P21 all males. Staining was used to identify neurons for quantitation of total hippocampal neurons, and regions CA1, CA2/3, and dentate gyrus. Maternal UN did not significantly alter neuronal cell number and distribution in any specific hippocampal region of P1 pups (Figure 2). However, in P21 pups there was a statistically significant decrease in neuronal number in the CA2/3 region of UN pups compared to controls (Figure 3).
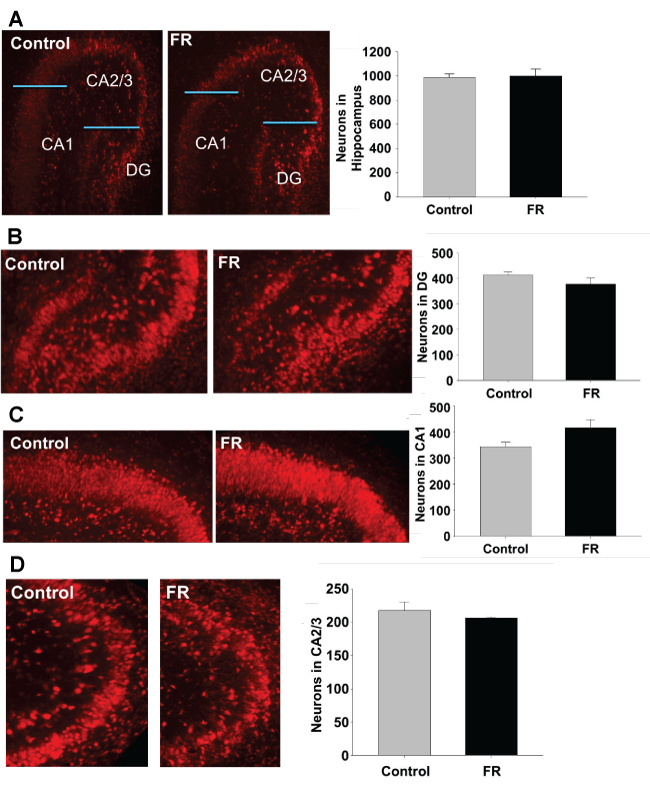
.
Figure 2: P1 hippocampus using NeuN to identify neurons. A to D represents immuno histo chemistry of P1 rat hippocampus showing pyramidal cells in combined regions, dentate gyrus, CA1, and CA2/3 respectively. There was no statistically significant difference between pyramidal cell numbers in UN pups compared to controls. All data are mean ± SEM (n = 3) *P < 0.05.
View Figure 2
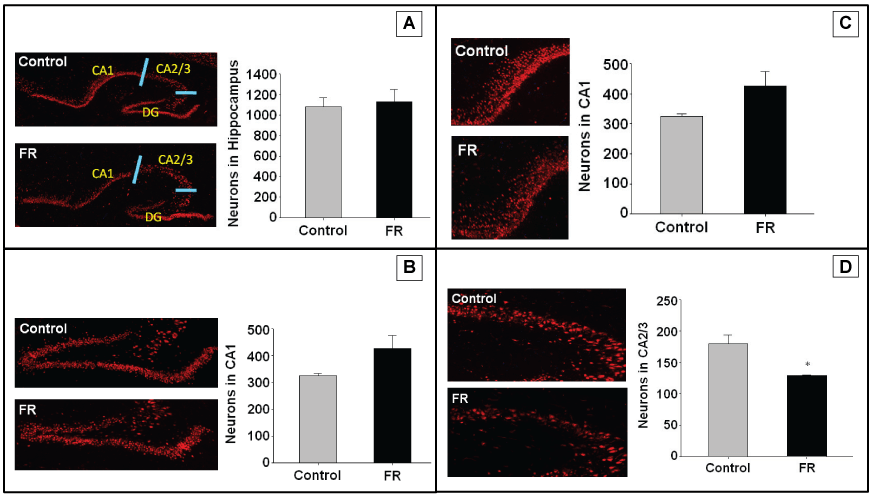
.
Figure 3: P21 hippocampus using NeuN to identify neurons. A to D represents immunohistochemistry of P1 rat hippocampus showing pyramidal cells in combined regions, dentate gyrus, CA1, and CA2/3 respectively. All data are mean ± SEM (n = 4) *P < 0.05.
View Figure 3
Statistical analysis
Identification of mRNA differential expression required that all the genes pass two conservative criteria: a differential expression of mRNA beyond the 95% confidence intervals compared to other homotypic genes. This 95% confidence interval corresponded to a 1.5 fold change in mRNA expression. The second criteria required that the differential compared between homotypic genes was a statistically significant difference of P < 0.01 using a paired t-test.
The Western blot data included mean values of neurogenin 1 and Notch 1 protein for control and UN pups. A two-tailed student’s t-test assuming unequal variances was used to determine a difference in the mean values between the groups (P < 0.05).
For immunohistochemistry, the total number of cells stained for NeuN in the hippocampus and each sub region (CA1, CA2/3, dentate gyrus) was manually counted and recorded, and the mean values calculated. A two-tailed student’s t-test assuming unequal variance was used to determine if there was a statistical difference in mean values between the groups (P < 0.05)
Discussion
Maternal undernutrition induced a definite effect on neuronal gene expression in the newborn rat brain. Our data shows that Ngn1 production (a neuronal progenitor determining factor) is down-regulated by nearly two-fold in maternal UN pups. These alterations in Ngn1 are reflected at both the genomic level (as demonstrated by microarray analysis) and at the cellular level (as seen in our Western blot expression).
To examine the mechanism for the decreased production of Ngn1, we explored the expression of Notch1, a factor implicated in partial control of Ngn1. Western blot expression demonstrated up regulation of Notch1 in the hippocampus of maternal UN pups compared to controls. These data suggest that decreased hippocampal neuronal cell expression of Ngn1 in response to maternal UN may be partially tied to the Notch pathway. Most importantly, when we examined actual hippocampal pyramidal cell quantification, we observed a decreased number of these cells in the CA2/3 region at P21. In sum, these data suggest a potential for altered neuronal function and brain development in response to maternal UN and offspring low birth weight.
In our study we focused our analysis on Ngn1, part of a super family of basic-helix-loop-helix proteins responsible for determination of neurologic cell fate [12,13], Other members of this family include Neurogenin 2 (Ngn2) and Neurogenin 3 (Ngn3) which are determining factors for epibranchial placode-derived sensory neurons and endocrine cell lines of the pancreas, respectively [14-16]. Ngn1 is expressed throughout the central and peripheral nervous systems. It is specifically found in the embryonic and prenatal hippocampus, proximal cranial nerves and sensory ganglion where it is essential for the determination of neuronal progenitor cells [15,17,18]. Our results clearly show that maternal UN-induced inhibition can be seen at various levels of Ngn1 production, from transcription to translation. As our microarray and Western blot analysis is of hippocampal tissues harvested from P1 rats, the resultant decreases in Ngn1 production are clearly due to intrauterine influences, specifically maternal and thus fetal nutrient restriction. We postulate that alterations in Notch1 pathway leads to down-regulation of Ngn1 in UN rat pups. Combined with evidence that intrauterine growth restriction is associated with reduced hippocampal volume in mice and reduced hippocampal neurons in guinea pigs [19,20], our findings of decreased neuronal number in the CA2/3 region of UN pups strongly suggests that low birth weight may result in altered hippocampal development.
The effects of food restriction are believed to be a diverse phenomenon affecting multiple organs and organ systems in fetal development [21-26]. The nervous system would not seem to be immune to these effects. Furthermore, there is growing support from epidemiologic studies that one of the components of the metabolic syndrome, specifically obesity, is directly linked to the development of dementia [27-29] The mechanistic connection between obesity and dementia is not fully understood, though it is generally accepted that obesity causes hyperinsulinemia, glucose intolerance and diabetes, all of which are risk factors for dementia [30]. These are also risk factors for peripheral vascular disease and potentially cerebral vascular disease which itself may have a direct action on the amyloid cascade increasing the progression of amyloid deposition [31]. Cerebral vascular disease may also work in conjunction with amyloid deposition by directly damaging brain tissue leaving the individual more susceptible to or with a decreased threshold for the development of dementia because of decreased neuronal reserve [32]. In fact, large vessel cerebral vascular disease increases the frequency of neuritic plaques seen in Alzheimer’s disease [33].
The decreased expression of tryptophan hydroxylase noted in our microarray analysis also poses an additional potential effect in neuronal development and intrauterine programmed predisposition to other adult neurologic disease, specifically affect disorders. Both anxiety-like and depressive-like behaviors are increased in the offspring of UN rats as exhibited by risk averse activities [34,35]. Morgese et al. showed that 5HT appears to be central to these processes in these offspring. Low synaptic 5HT levels are seen in patients that exhibit anxiety and depression disorders. Increasing the synaptic levels of 5HT, as with selective serotonin reuptake inhibiting drugs alleviate these symptoms and are well established management strategies of these conditions in humans. As tryptophan hydroxylase is the enzymatic rate limiting step in 5HT synthesis, it would follow that low levels of the enzyme as seen in our microarray analysis in our UN pups may predispose to these offspring to anxiety-like and depressive-like disorders as seen in humans
With regards to our Ngn1 and Notch data, despite this widely accepted pathway, our results suggest an additional mechanism linking obesity and Alzheimer’s disease, with both attributable in part to the fetal nutrient environment. Our findings of maternal UN-induced down-regulation of Ngn1 through Notch1 inhibition is an important step towards possible understanding of pathologic neurologic development and a potential means of prevention through proper maternal nutrition throughout pregnancy.
Acknowledgement
This work was supported by the National Institutes of Health Grants R01HD054751 and R01DK081756 and Los Angeles Biomedical Research Institute (SLK).
Conflict of Interest
The authors report no conflict of interest.
References
-
Hanson M (2015) The birth and future health of DOHaD. J Dev Orig Health Dis 6: 434-437.
-
Barker DJ, Osmond C (1986) Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet 1: 1077-1081.
-
Desai M, Li T, Ross MG (2011) Hypothalamic neurosphere progenitor cells in low birth-weight rat newborns: Neurotrophic effects of leptin and insulin. Brain Res 1378: 29-42.
-
Desai M, Li T, Han G, Ross MG (2014) Programmed hyperphagia secondary to increased hypothalamic SIRT1. Brain Res 1589: 26-36.
-
Desai M, Gayle D, Babu J, Ross MG (2005) Programmed obesity in intrauterine growth-restricted newborns: modulation by newborn nutrition. Am J Physiol Regul Integr Comp Physiol 288: R91-R96.
-
Karsten SL, Sang TK, Gehman LT, Chatterjee S, Liu J, et al. (2006) A genomic screen for modifiers of tauopathy identifies puromycin-sensitive aminopeptidase as an inhibitor of tau-induced neurodegeneration. Neuron 51: 549-560.
-
Kudo LC, Parfenova L, Vi N, Lau K, Pomakian J, et al. (2010) Integrative gene-tissue microarray-based approach for identification of human disease biomarkers: application to amyotrophic lateral sclerosis. Hum Mol Genet 19: 3233-3253.
-
Kudo LC, Parfenova L, Ren G, Vi N, Hui M, et al. (2011) Puromycin-sensitive aminopeptidase (PSA/NPEPPS) impedes development of neuropathology in hPSA/TAU(P301L) double-transgenic mice. Hum Mol Genet 20: 1820-1833.
-
Ohmi K, Kudo LC, Ryazantsev S, Zhao HZ, Karsten SL, et al. (2009) Sanfilippo syndrome type B, a lysosomal storage disease, is also a tauopathy. Proc Natl Acad Sci U S A 106: 8332-8337.
-
Hiroi R, Carbone DL, Zuloaga DG, Bimonte-Nelson HA, Handa RJ (2016) Sex-dependent programming effects of prenatal glucocorticoid treatment on the developing serotonin system and stress-related behaviors in adulthood. Neurosci 320: 43-56.
-
Ma Q, Kintner C, Anderson DJ (1996) Identification of neurogenin, a vertebrate neuronal determination gene. Cell 87: 43-52.
-
Pozzoli O, Bosetti A, Croci L, Consalez GG, Vetter ML (2001) Xebf3 is a regulator of neuronal differentiation during primary neurogenesis in Xenopus. Dev Biol 233: 495-512.
-
Sommer L, Ma Q, Anderson DJ (1996) Neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol Cell Neurosci 8: 221-241.
-
Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, et al. (1998) The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron 20: 483-494.
-
Ma Q, Fode C, Guillemot F, Anderson DJ (1999) Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev 13: 1717-1728.
-
Gradwohl G, Dierich A, LeMeur M, Guillemot F (2000) Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A 97: 1607-1611.
-
Ma Q, Chen Z, del BB I, de la Pompa JL, Anderson DJ (1998) Neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron 20: 469-482.
-
Seo S, Lim JW, Yellajoshyula D, Chang LW, Kroll KL (2007) Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J 26: 5093-5108.
-
Mallard EC, Rehn A, Rees S, Tolcos M, Copolov D (1999) Ventriculomegaly and reduced hippocampal volume following intrauterine growth-restriction: implications for the aetiology of schizophrenia. Schizophr Res 40: 11-21.
-
Mallard C, Loeliger M, Copolov D, Rees S (2000) Reduced number of neurons in the hippocampus and the cerebellum in the postnatal guinea-pig following intrauterine growth-restriction. Neurosci 100: 327-333.
-
Eriksson JG, Forsén T, Tuomilehto J, Osmond C, Barker DJ (2001) Early growth and coronary heart disease in later life: longitudinal study. BMJ 322: 949-953.
-
Eriksson JG, Forsén T, Tuomilehto J, Osmond C, Barker DJ (2003) Early adiposity rebound in childhood and risk of Type 2 diabetes in adult life. Diabetologia 46: 190-194.
-
Forsén T, Osmond C, Eriksson JG, Barker DJ (2004) Growth of girls who later develop coronary heart disease. Heart 90: 20-24.
-
Frankel S, Elwood P, Sweetnam P, Yarnell J, Smith GD (1996) Birth weight, body-mass index in middle age, and incident coronary heart disease. Lancet 348: 1478-1480.
-
Gesta S, Blüher M, Yamamoto Y, Norris AW, Berndt J, et al. (2006) Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc Natl Acad Sci U S A 103: 6676-6681.
-
Stein CE, Fall CH, Kumaran K, Osmond C, Cox V, et al. (1996) Fetal growth and coronary heart disease in south India. Lancet 348: 1269-1273.
-
Luchsinger JA, Gustafson DR (2009) Adiposity, type 2 diabetes, and Alzheimer's disease. J Alzheimers Dis 16: 693-704.
-
Milionis HJ, Florentin M, Giannopoulos S (2008) Metabolic syndrome and Alzheimer's disease: a link to a vascular hypothesis? CNS Spectr 13: 606-613.
-
Miller DB, O'Callaghan JP (2008) Do early-life insults contribute to the late-life development of Parkinson and Alzheimer diseases? Metabolism 57 Suppl 2: S44-49.
-
Luchsinger JA, Mayeux R (2004) Cardiovascular risk factors and Alzheimer's disease. Curr Atheroscler Rep 6: 261-266.
-
Honig LS, Scarmeas N (2005) Most cases of dementia with hippocampal sclerosis may represent frontotemporal dementia. Neurology 64: 1102.
-
Luchsinger JA, Mayeux R (2007) Adiposity and Alzheimer's disease. Curr Alzheimer Res 4: 127-134.
-
Cummings JL (2004) Alzheimer's disease. N Engl J Med 351: 56-67.
-
Levay EA, Paolini AG, Govic A, Hazi A, Penman J, Kent S (2008) Anxiety-like behaviour in adult rats perinatally exposed to maternal calorie restriction. Behav Brain Res 191: 164-172.
-
Morgese MG, Tucci P, Mhillaj E, Bove M, Schiavone S, et al. (2016) Lifelong Nutritional Omega-3 Deficiency Evokes Depressive-Like State Through Soluble Beta Amyloid. Mol Neurobiol.





