International Journal of Pathology and Clinical Research
Liquid Biopsy: A Future Tool for Post Treatment Surveillance in Head and Neck Cancer?
Joost H Van Ginkel1,2, Manon MH Huibers2, Rob Noorlag1, Remco de Bree3, Robert JJ van Es1,3 and Stefan M Willems2*
1Department of Oral and Maxillofacial Surgery, University Medical Center Utrecht, the Netherlands
2Department of Pathology, University Medical Center Utrecht, the Netherlands
3Department of Head and Neck Surgical Oncology, UMC Utrecht Cancer Center, University Medical Center Utrecht, Utrecht, the Netherlands
*Corresponding author:
Stefan M Willems, MD, PhD, Department of Pathology, Heidelberglaan 100, 3581 CX Utrecht, The Netherlands, E-mail: s.m.willems-4@umcutrecht.nl
Int J Pathol Clin Res, IJPCR-1-013, (Volume 1, Issue 1), Research Article; ISSN: 2469-5807
Received: October 01, 2015 | Accepted: November 25, 2015 | Published: November 30, 2015
Citation: Van Ginkel JH, Huibers MMH, Noorlag R, de Bree R, van Es RJJ, et al. (2015) Liquid Biopsy: A Future Tool for Post Treatment Surveillance in Head and Neck Cancer?. Int J Pathol Clin Res 1:013. 10.23937/2469-5807/1510013
Copyright: © Van Ginkel JH, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
The prognosis of head and neck squamous cell carcinoma (HNSCC) is largely based on disease stage. Despite improvements in treatment, recurrence rates are still considered high. Currently, disease progression or regression after curative treatment is monitored by clinical evaluation combined with flexible endoscopy and/or imaging. However, specificity of imaging is low due to post treatment effects. Detection of circulating tumor DNA (ctDNA) from blood samples of HNSCC patients is a minimally invasive technique that could lead to an earlier detection of recurrences. In addition, digital droplet PCR (ddPCR) could be used to sensitively detect these mutational targets. Future studies on ctDNA using ddPCR in blood samples of HNSCC patients is recommended during the follow up stage to detect recurrences in a timely manner.
Keywords
Head neck cancer, Surveillance, Liquid biopsy, Digital polymerase chain reaction
Introduction
Head and neck squamous cell carcinomas (HNSCCs) comprise tumors of the oral cavity, oropharynx, hypopharynx and larynx. The prognosis of HNSCC is largely based on the disease stage at presentation, particularly the presence of lymph node metastases in the neck and distant metastases. Despite improvements in treatment, locoregional recurrence rates range from 25% to 50% depending on tumor location and stage. In case of recurrence above the clavicles, salvage surgery remains the only curative option [1]. Therefore, timely diagnosis of locoregional recurrence is crucial to increase the possibility of prompt curative salvage surgery [2]. To date, the standard method for assessment of locoregional control is clinical evaluation supported by flexible endoscopy and/or imaging. Therefore, post treatment imaging such as computed tomography (CT), magnetic resonance imaging (MRI) or F-18-fluorodeoxyglucose positron emission tomography (FDG-PET) in case of clinical suspicion of recurrence is performed promptly. However, differentiation between post treatment effects and tumor recurrence in these imaging modalities is difficult, especially after (chemo) radiotherapy. Although very sensitive (94%), the specificity of the FDG-PET for locoregional recurrence is lower (82%), alongside a positive predictive value of 75% as described in a systematic review and meta-analysis of PET trials for detecting residual or recurrent HNSCC treated by (chemo)radiotherapy [3]. This could lead to a considerable amount of false positive cases rendering erroneous therapeutic interpretation. As a result, there is a clear need for more specific markers to detect (early) recurrences. With the discovery of the genetic aberrations that induce the initiation and progression of malignant tumors, detection of specific mutations are increasingly being used as biomarkers to predict local recurrence. Moreover, detection of tumor DNA, either as freely circulating tumor DNA (ctDNA) or in circulating tumor cells (CTCs), has proven to be a promising tool to detect early spread of cancer cells in blood [4,5]. More recently, these so-called liquid biopsies have already shown promising results as genetic biomarkers to predict early recurrences in different cancer types [5-7].
In this paper we will shortly discuss the various advantages, drawbacks and current clinical applications of liquid biopsy in oncology, as well as its use as a novel predictive tool in the post treatment surveillance in HNSCC patients. Furthermore, we will delineate the various advantages of a novel PCR technique, the droplet digital polymerase chain reaction (ddPCR) and the potential implementation of liquid biopsy and ddPCR into daily clinical practice in the future. We will support the latter by offering a possible workflow of our strategy.
HNSCC and TP53
The pathogenesis of HNSCC is strongly related to alcohol consumption, tobacco use and high-risk human papilloma virus (HPV) infection [8]. HPV-negative and HPV-positive tumors have been shown to be (epi) genetically and clinically different entities [9]. Chung et al. found PIK3CA and PTEN in HPV-positive tumors and TP53 (87%) and CDKN2A/B (54%) in HPV-negative tumors to be the most frequently altered genes in a cohort of 252 HNSCC patients [10]. Moreover, TP53 mutations are the most common genetic aberrations in HPV-negative HNSCC and have an early onset in the carcinogenesis of HNSCCs [11-14]. Subsequent loss of heterozygosity (LOH) leads to loss of the non-mutated gene [15] and newly formed clonal expansions [16,17]. Specific TP53 mutations in the clonal progenitor and expansion cells could be used as a biomarker to detect the presence of primary and/or metastatic tumor.
Liquid Biopsy
Tissue biopsy in conjunction with PCR sequencing techniques is already applied in clinical practice in order to guide cancer treatment modalities. However, major drawbacks of tissue biopsy are biased acquisition of tissue due to intratumoral heterogeneity and, risk of complications, due to its invasiveness [18]. Moreover, HNSCC locoregional (micro) metastases are often too small to be sampled and detected by tissue biopsy sampling. Consequently, although tissue biopsies can deliver useful information about the primary tumor genetic profile, there are limitations to its use as a method for monitoring post treatment locoregional disease surveillance.
Blood samples are another source to retrieve DNA (and RNA) in an attempt to monitor tumor status. First identified in 1948, blood of healthy individuals contains genetic material in the form of cell free DNA (cfDNA) [19]. Additionally, in patients with a malignant tumor, blood can contain CTCs [20-22], probably being released into the bloodstream by the (metastatic) tumor cells following apoptosis and necrosis, or by active release of living cells [23]. CfDNA is already examined clinically in prenatal testing, transplant patients and trauma patients [24,25], while ctDNA is found to play a role as a potential prognostic tool in cancer treatment [26,27].
The clinical applications of ctDNA in cancer treatment are roughly divided into two categories: characterizing tumor genetics and quantitation of ctDNA representing tumor burden [28]. Applications based on ctDNA for characterizing tumor genetics can be used as a tool to guide targeted drug therapy, particularly in metastatic disease and as an alternative to conventional tissue biopsy in cases of absolute or relative contraindications to biopsy. The second category of applications could be used as a disease monitoring tool, as there appears to be a relation between ctDNA load and tumor burden [29].
Diehl et al. evidenced that in many patients who underwent curative colorectal cancer surgery and developed disease recurrence, plasma ctDNA with tumor specific genetic alterations was still detectable after surgery [30]. In another pilot study the possibility of prognostication and monitoring of oral squamous cell carcinoma was assessed by analyzing serum-isolated DNA of 64 patients using microsatellite markers to detect allelic imbalances. In more than 50% of patients allelic imbalances were identified in serum DNA corresponding with tumor DNA. In turn, a correlation was found between allelic imbalances in serum DNA and tumor stage [27]. Although the uses of microsatellites have been subject of debate [31], these results are promising. More recently, Bettegowda et al. reported detectable ctDNA levels in 55% of 223 patients with localized tumors of varying origin (i.e. pancreatic, ovarian, colorectal, bladder, gastro-esophageal, breast, melanoma, hepatocellular and head and neck cancers). Also, a directly proportional correlation was found between the fraction of patients with detectable ctDNA levels and tumor stage [32].
Similarly as ctDNA, CTCs could be used to genetically analyze tumor DNA for targeted treatment strategies and to detect a recurrence or progression of disease after initial therapy. The main principle of this technique relies on the identification of CTCs by enrichment of cells expressing epithelial cell adhesion molecules followed by immunofluorescent staining using different markers.
In a pooled analysis of 1944 patients with metastatic breast cancer, an independent prognostic effect of CTC enumeration on progression-free survival and overall survival was confirmed [33]. All patients were analyzed at the start of a new therapy line. Patients with a CTC count of 5 per 7.5 ml or higher at the start of treatment were associated with a decreased progression-free survival and overall survival (p < 0.0001), compared to patients with a CTC count of less than 5 per 7.5 ml. Another field of interest in current research is CTC characterization. Several studies investigated protein expression, RNA expression and DNA aberrations in CTCs to help guide drug therapy in different cancer types. Especially HER2 protein expression in breast cancer patients and AR signaling in castration resistant prostate cancer are subjects of interest in current studies [34]. There are however, certain issues pertaining to the use of CTCs before translating into the therapeutic arena, particularly with regards to detection and characterization of the genetic alterations, because CTC concentration in blood circulation is very low. Current estimations on CTC concentration is one tumor cell per one billion of normal blood cells. A sensitive way to detect, isolate and differentiate CTCs remains an underlying challenge. The detection of CTCs is also dependent on the time point of blood sampling. In HNSCC studies, the portion of patients with positive CTC levels varies from 6.5 to 87.5% [21,35-37].
Regarding post treatment tumor monitoring, liquid biopsy could be a valuable tool in the follow-up of HNSCC patients. Tumor specific genetic alterations can be detected by analyzing ctDNA with mutation specific primer assays. These latter are custom selected and based on data from whole-exome sequencing (WES) performed on HNSCC [13,14]. By using next-generation sequencing (NGS) on a gene panel based platform, tumor or patient specific mutations for each HNSCC will be determined. Using a more targeted approach instead of WES allows a more sensitive sequencing method for primary tumors [38,39]. By taking liquid biopsies from the patient at different time points during follow-up after (chemo) radiotherapy or resection of the primary tumor, ctDNA can be isolated and used as a biomarker based on information of previously performed NGS. Due to the relatively short half-life of ctDNA of approximately 2 hours, tumor changes can be evaluated in hours rather than weeks to months [18]. This allows to monitor disease progression or regression very closely and the early detection of tumor recurrence or metastases after initial treatment with curative intent. This manner of personalized cancer management has the potential to prevent overtreatment and insufficient treatment with the possibility to avoid complications of invasive diagnostic techniques and disease progression respectively.
Although the earlier mentioned data underscore the emerging evidence that monitoring genetic alterations in ctDNA is a promising tool to monitor disease recurrence and stage, several important technical and biological obstacles has yet to be removed in order to be able to implement this diagnostic tool in clinical practice [28]. Firstly, analytic sensitivity is necessary to reliably isolate and detect ctDNA when present in the blood. Secondly, a high proportion of patients should carry detectable amounts of ctDNA, because absolute ctDNA levels vary within each subpopulation [32]. Thirdly, a low signal-to-noise ratio due to the presence of high levels of cfDNA (i.e. wild type DNA) could interfere with the detection of target ctDNA. Lastly, tumor heterogeneity remains an important challenge. Clonal expansions can arise within the primary tumor, carrying a different mutational profile. During tissue biopsy for initial genetic tumor profiling, a (rare) subclone could be selected to detect ctDNA. This could possibly lead to inaccurate representation of tumor burden due to low levels of targeted ctDNA [27].
Droplet Digital PCR
A novel tool in genetic diagnostics is ddPCR. DNA samples will be analyzed using specific prefabricated probes. These assays are mutation specific and correlate with tumor mutations previously found in corresponding tissue biopsies. Combined with PCR specific primers, samples will be processed with oil to create 20,000 water-in-oil droplets containing the DNA molecules. The readout of the droplets is an end-point PCR based on Poisson statistics, suggesting that target DNA molecules are distributed randomly to the droplets. After actual PCR and amplification has been conducted, some reactions contain target copies while others do not. The yield will be a read out as positive end-point and negative end-point respectively [40,41]. This enables the absolute quantitation of nucleic acids in a sample, which will provide great precision due to the partition of the sample by 20,000 fold. Thus, this will facilitate the detection of rare targets of interest. Furthermore, accurate quantification of targets inside the droplets enables the reduction of error rates due to normal PCR efficiency bias. This could accommodate the earlier mentioned issue of a low signal-to-noise ratio in ctDNA detection. Although (prospective) data regarding the sensibility of ddPCR in this setting are currently lacking, these advantages of ddPCR might efficiently contribute to the diagnostic process in post-treatment monitoring of HNSCC patients.
Future Directives
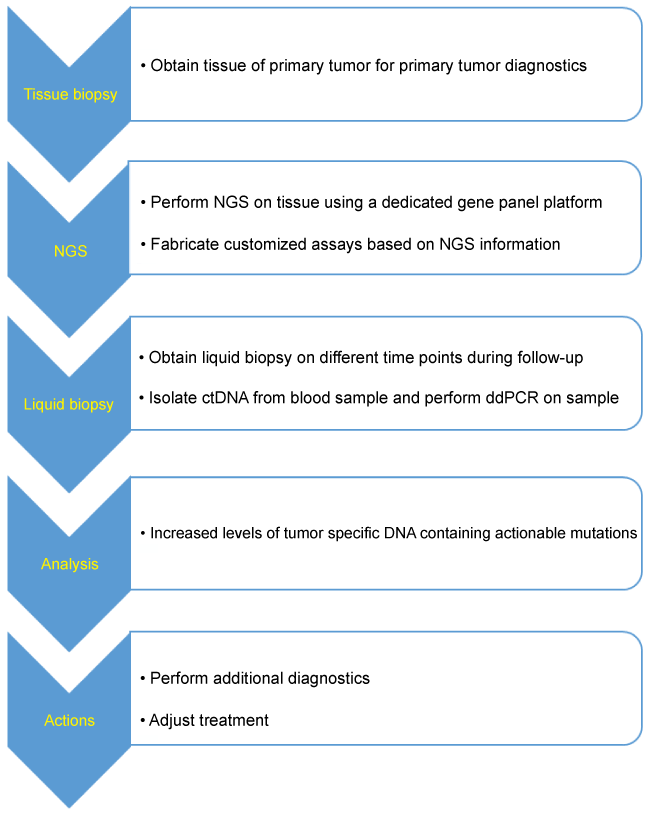
Our future goals are to assess the role of liquid biopsy in the locoregional surveillance of HNSCC patients following curative treatment and to determine if ddPCR is a sensitive and specific PCR technique for the detection of tumor specific alterations in ctDNA. In order to do so, pilot experiments will be carried out to see if this technique is feasible for the proposed aim. A prospective longitudinal study of HNSCC patients, will allow for the analysis of blood samples at different time points: before treatment, after treatment and at recurrence. These future studies will attempt to determine the feasibility to use minimally invasive and sensitive techniques for HNSCC patients disease surveillance after treatment and set up a structuralized workflow for the use of liquid biopsy in clinical practice (Figure 1).
Conflict of Interest
The authors declare no conflicts of interest.
Authors' contributions
Joost H. van Ginkel and M. Huibers contributed equally
Acknowledgement
The authors would like to thank R de Weger for setting up the technique of the ddPCR, AH Gijsbers-Bruggink for her help with acquiring biomaterials and E Siera-de Koning and R Wiggers for their excellent help with using the ddPCR and setting up and performing experiments so far.
References
-
Ho AS, Kraus DH, Ganly I, Lee NY, Shah JP, et al. (2014) Decision making in the management of recurrent head and neck cancer. Head Neck 36: 144-151.
-
de Bree R, van der Putten L, Brouwer J, Castelijns JA, Hoekstra OS, et al. (2009) Detection of locoregional recurrent head and neck cancer after (chemo)radiotherapy using modern imaging. Oral Oncol 45: 386-393.
-
Isles MG, McConkey C, Mehanna HM (2008) A systematic review and meta-analysis of the role of positron emission tomography in the follow up of head and neck squamous cell carcinoma following radiotherapy or chemoradiotherapy. Clin Otolaryngol 33: 210-222.
-
da Silva Filho BF, Gurgel AP, Neto MA, de Azevedo DA, de Freitas AC, et al. (2013) Circulating cell-free DNA in serum as a biomarker of colorectal cancer. J Clin Pathol 66: 775-778.
-
Hao TB, Shi W, Shen XJ, Qi J, Wu XH, et al. (2014) Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br J Cancer 111: 1482-1489.
-
Umetani N, Giuliano AE, Hiramatsu SH, Amersi F, Nakagawa T, et al. (2006) Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol 24: 4270-4276.
-
Qiu M, Wang J, Xu Y, Ding X, Li M, et al. (2015) Circulating tumor DNA is effective for the detection of EGFR mutation in non-small cell lung cancer: a meta-analysis. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 24: 206-212.
-
Leemans CR, Braakhuis BJ, Brakenhoff RH (2011) The molecular biology of head and neck cancer. Nat Rev Cancer 11: 9-22.
-
van Kempen PM, Noorlag R, Braunius WW, Moelans CB, Rifi W, et al. (2015) Clinical relevance of copy number profiling in oral and oropharyngeal squamous cell carcinoma. Cancer Med 4: 1525-1535.
-
Chung CH, Guthrie VB, Masica DL, Tokheim C, Kang H, et al. (2015) Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Ann Oncol 26: 1216-1223.
-
Nees M, Homann N, Discher H andl T, Enders C, et al. (1993) Expression of Mutated p53 Occurs in Tumor-distant Epithelia of Head and Neck Cancer Patients: A Possible Molecular Basis for the Development of Multiple Tumors. Cancer Res 53: 4189-4196
-
Soder AI, Hopman AH, Ramaekers FC, Conradt C, Bosch FX (1995) Distinct nonrandom patterns of chromosomal aberrations in the progression of squamous cell carcinomas of the head and neck. Cancer Res 55: 5030-5037.
-
Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, et al. (2011) Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333: 1154-1157.
-
Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, et al. (2011) The mutational landscape of head and neck squamous cell carcinoma. Science 333: 1157-1160.
-
Graveland AP, Golusinski PJ, Buijze M, Douma R, Sons N, et al. (2011) Loss of heterozygosity at 9p and p53 immunopositivity in surgical margins predict local relapse in head and neck squamous cell carcinoma. Int J Cancer 128: 1852-1859.
-
Califano J, van der Riet P, Westra W, Nawroz H, Clayman G, et al. (1996) Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res 56: 2488-2492.
-
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100: 57-70.
-
Diaz LA Jr, Bardelli A (2014) Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 32: 579-586.
-
Mandel P, Metais P (1948) Les acides nucleiques du plasma sanguin chez l'homme. C R Seances Soc Biol Fil 142: 241-243.
-
Alix-Panabieres C, Schwarzenbach H, Pantel K (2012) Circulating tumor cells and circulating tumor DNA. Annu Rev Med 63: 199-215.
-
Pantel K, Brakenhoff RH, Brandt B (2008) Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer 8: 329-340.
-
Wollenberg B, Walz A, Kolbow K, Pauli C, Chaubal S, et al. (2004) Clinical relevance of circulating tumour cells in the bone marrow of patients with SCCHN. Onkologie 27: 358-362.
-
Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A (2013) Liquid biopsy: monitoring cancer-genics in the blood. Nat Rev Clin Oncol 10: 472-484.
-
Lui YY, Dennis YM (2002) Circulating DNA in plasma and serum: biology, preanalytical issues and diagnostic applications. Clin Chem Lab Med 40: 962-968.
-
Lo YM, Rainer TH, Chan LY, Hjelm NM, Cocks RA (2000) Plasma DNA as a prognostic marker in trauma patients. Clin Chem 46: 319-323.
-
Chen Z, Fadiel A, Naftolin F, Eichenbaum KD, Xia Y (2005) Circulation DNA: biological implications for cancer metastasis and immunology. Med Hypotheses 65: 956-961.
-
Hamana K, Uzawa K, Ogawara K, Shiiba M, Bukawa H, et al. (2005) Monitoring of circulating tumour-associated DNA as a prognostic tool for oral squamous cell carcinoma. Br J Cancer 92: 2181-2184.
-
Ignatiadis M, Lee M, Jeffrey SS3 (2015) Circulating Tumor Cells and Circulating Tumor DNA: Challenges and Opportunities on the Path to Clinical Utility. Clin Cancer Res 21: 4786-4800.
-
Thierry AR, Mouliere F, Gongora C, Ollier J, Robert B, et al. (2010) Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res 38: 6159-6175.
-
Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, et al. (2008) Circulating mutant DNA to assess tumor dynamics. Nat Med 14: 985-990.
-
Fleischhacker M, Schmidt B (2007) Circulating nucleic acids (CNAs) and cancer--a survey. Biochim Biophys Acta 1775: 181-232.
-
Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, et al. (2014) Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6: 224ra24.
-
Bidard FC, Peeters DJ, Fehm T, Nole F, Gisbert-Criado R, et al. (2014) Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol 15: 406-414.
-
Jiang Y, Palma JF, Agus DB, Wang Y, Gross ME (2010) Detection of androgen receptor mutations in circulating tumor cells in castration-resistant prostate cancer. Clin Chem 56: 1492-1495.
-
Ignatiadis M, Dawson SJ (2014) Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Ann Oncol 25: 2304-2313.
-
Mockelmann N, Laban S, Pantel K, Knecht R (2014) Circulating tumor cells in head and neck cancer: clinical impact in diagnosis and follow-up. Eur Arch Otorhinolaryngol 271: 15-21.
-
Pantel K, Speicher MR (2015) The biology of circulating tumor cells. Oncogene: 2015 Jun 8.
-
Gagan J, Van Allen EM (2015) Next-generation sequencing to guide cancer therapy. Genome Med 7: 80.
-
Jones S, Anagnostou V, Lytle K, Parpart-Li S, Nesselbush M, et al. (2015) Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med 7: 283ra53.
-
Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, et al. (2011) High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 83: 8604-8610.
-
Zec H, Shin DJ, Wang TH (2014) Novel droplet platforms for the detection of disease biomarkers. Expert Rev Mol Diagn 14: 787-801.