International Journal of Stem cell Research and Therapy
Function of Sirtuins in Cancer Stem Cells
Zhen Dong and Hongjuan Cui*
State Key Laboratory of Silkworm Genome Biology, Southwest University, P.R. China
*Corresponding author: Prof. Hongjuan Cui, Department of Stem Cell Biology and Translational Medicine, No.02, Tiansheng Road, Beibei District, Chongqing, China, Tel: +86-023-68251713, Fax: +86-023-68251128, E-mail: hongjuan.cui@gmail.com, hcui@swu.edu.cn
Int J Stem Cell Res Ther, IJSCRT-3-024, (Volume 3, Issue 1), Review Article; ISSN: 2469-570X
Received: December 14, 2015 | Accepted: February 05, 2016 | Published: February 08, 2016
Citation: Dong Z, Cui H (2016) Function of Sirtuins in Cancer Stem Cells. Int J Stem Cell Res Ther 3:024. 10.23937/2469-570X/1410024
Copyright: © 2016 Dong Z, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Cancer stem cells (CSCs) are self-renewing cancer cells in tumors that are suggested to be responsible for tumor initiation, progression and relapse. The molecular mechanisms of CSCs biology could provide some novel managements for cancer treatment. Sirtuins, as a family of class III deacetylases, are recently found to play essential roles in the maintenance and differentiation of various cancer stem cells, including glioma stem cells (GSCs), colorectal cancer stem cells (CCSCs), breast cancer stem cells (BCSCs), pancreatic cancer stem cells (PCSCs), chronic myeloid leukemia (CML) and acute myeloid leukemia (AML) stem cells. Herein, we reviewed the advanced studies that linking sirtuins, especially SIRT1 and SIRT2, to cancer stem cells and will also discuss how these enzymes could be used in biological pharmaceutics of tumors.
Keywords
Sirtuins, Cancer stem cells, Epigenetic regulation, Pharmaceutics
Abbreviations
ABCB5: ATP-binding Cassette B5, ADP: Adenosine Diphosphate, ALDH1: Aldehyde Dehydrogenase 1, ALDH1A1: Aldehyde Dehydrogenase 1A1, ALL: Acute Lymphoblastic Leukemia, AML: Acute Myeloid Leukemia, BAX: Bcl2-Associated X Protein, BCR-ABL: Breakpoint Cluster Region-C-Abl Oncogene 1, BCSC: Breast Cancer Stem Cell, CCSC: Colorectal Cancer Stem Cell, CD24: Cluster of Differentiation 24, CD44: Cluster of Differentiation 44, CD90: Cluster of Differentiation 90, CD133: Cluster of Differentiation 133, CD200: Cluster of Differentiation 200, CDK6: cyclin-dependent kinase 6, CTE: C-Terminal Extension, CLL: Chronic Lymphocytic Leukemia, CML: Chronic Myeloid Leukemia, CNSC: Cancerous Neural Stem Cell, CPEB1: Cytoplasmic Polyadenylation Element Binding Protein 1, CSC: Cancer Stem Cell, EpCAM: Epithelial Cell Adhesion Molecule, DNA: Deoxyribonucleic Acid, ESA: Epithelial Specific Antigen, FLT3-ITD: Fms-Like Tyrosine Kinase 3 Internal Tandem Duplication, GFAP: Glial Fibrillary Acidic Protein, GSC: Glioma Stem Cell, HAT: Histone Acetyltransferase, HCAM, Homing Cell Adhesion Molecule, HDAC: Histone Deacetylase, HIF-1α: Hypoxia Inducible Factor 1α, HSA, Heat Stable Antigen, HSC: Hematopoietic Stem Cell, LSC: Leukemia Stem Cells, MPT: Mitochondrial Permeability Transition, NAD: Nicotinamide Adenine Dinucleotide, NF-κB: Nuclear Factor-κβ, NICD: Notch Intracellular Domain, NTE: N-Terminal Extension, OX2G: OX-2 Membrane Glycoprotein, PCSC: Pancreatic Cancer Stem Cell, PROM1: Prominin-1, ROS: Reactive Oxygen Species, Sox2: Sex Determining Region Y-box 2, SP: Side Population, TERT: Telomerase Reverse Transcriptase, THY1: Thy-1 Cell Surface Antigen, TKI: Tyrosine Kinase Inhibitor, USP22: Ubiquitin Specific Peptidase 22.
Short Review
In common tumor theories, tumor is an abnormal somatic cell proliferation caused by accumulation of random mutations in essential genes that control cell growth. However, the mutation theory have been challenged by various contradictions [1], such as oncogene activation appearance in normal cells, a slower growth rate of some tumor cells than that of normal cells, normal tissue development by injecting malignant teratoma cells into normal blastaea, silent metastatic cancers, tumor occurrence via transplanting normal tissue into the spleen, and outpouring relapse of cancers [2]. More importantly, minor subpopulations of cancer cells have aberrant self-renewal capacities to differentiate into several lineages of cancer cells. These self-renewing cancer cells are called cancer stem cells (CSCs), which are supposedly responsible for tumorigenesis, tumor spread, tumor resistance and tumor relapse [3].
As so far, CSCs can be identified by some cell surface markers such as cluster of differentiation 24 (CD24, or heat stable antigen CD24, HSA), cluster of differentiation 44 (CD44, also referred to as homing cell adhesion molecule, HCAM), cluster of differentiation 90 (CD90, also known as THY1), cluster of differentiation 133 (CD133, also known as prominin-1, PROM1), cluster of differentiation 200 (CD200, also called OX-2 membrane glycoprotein, OX2G), epithelial specific antigen (ESA, also called epithelial cell adhesion molecule, EpCAM), ATP-binding cassette B5 (ABCB5), nestin and so on [4]. The CSCs hypothesis provides a reasonable cellular mechanism to explain the therapeutic tolerances and dute behaviors exhibited in tumors, and CSCs has been suggested to be a significant barrier for efficient cancer therapy.
Recently, accumulative evidences showed that epigenetic mechanisms were key regulators during the process of differentiation from stem cells to specialized cells. In the development of tumors, aberrant epigenetic changes may help cancer cells to up or downregulate cancer stem cell markers in order to give rise a heterogeneous population of tumor cells [5]. Therefore, epigenetic regulation in cancer stem cells may provide new therapy for cancer treatment. Among them, histone acetylation is an important gene transcriptional regulation, which conducted by histone acetyltransferases (HATs) and histone deacetylases (HDACs). HDACs are grouped into 4 classes including class I, II, III and IV [6].
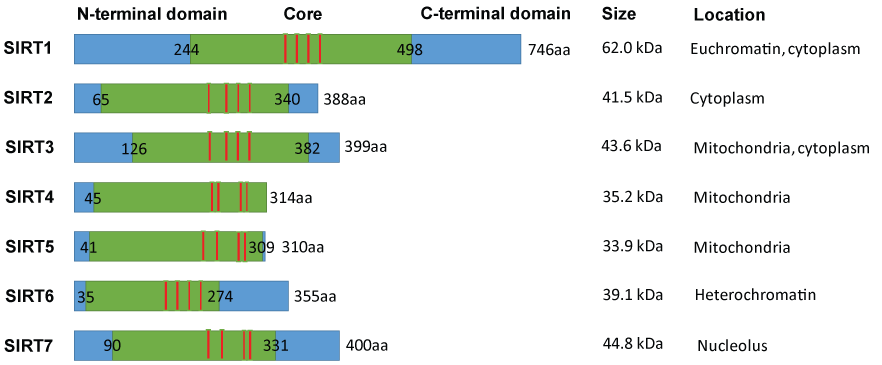
Class III HDACs, also called sirtuins, are a family of proteins composed of 7 members including SIRT1-7 [7]. As shown in figure 1, they are nicotinamide adenine dinucleotide (NAD+) dependent deacetylases and/or mono-[Adenosine diphosphate (ADP)-ribosyl] transferases that contain an N-terminal extension (NTE), a C-terminal extension (CTE), an NAD binding domain and a zinc-binding domain [8-10]. And they have different localizations: SIRT1/6/7 are localized in nucleus; SIRT1/2/3 are localized in cytoplasm; while SIRT3/4/5 are localized in mitochondria [10,11]. Sirtuins participated in multiple biological functions including chromatin structure and gene expression regulation, metabolic homeostasis, programmed cell death, aging, telomere activity, DNA repair, and cell differentiation [12]. Recently, more and more evidences showed that sirtuins, especially SIRT1 and SIRT2, played essential roles in the maintenance and differentiation of various cancer stem cells.
.
Figure 1: Schematic overview of Homo. Sapiens sirtuins family members SIRT1–7, NAD-binding poket (green), zinc-binding domain (red), and their size and intracellular locations.
View Figure 1
As the most important member in sirtuins, SIRT1 regulates multiple cellular processes including DNA repair, aging, metabolism, cell cycle, and survival [13] and plays an important role in maintaining self-renewal and differentiation of embryonic stem cells and hematopoietic stem cells (HSCs), especially under conditions of outside stress [14]. SIRT1 is a controversial histone deacetylase in tumorigenesis as it deacetylates both oncogenes such as β-catenin, survivin, and nuclear factor-κβ (NF-κβ) and tumor suppressors such as p53 and fork-head class O (FOXO) transcription factor [15]. But recently studies showed that SIRT1 was highly expressed in cancer stem cells, including glioma stem cells (GSCs) [16], colorectal cancer stem cells (CCSCs) [17], breast cancer stem cells (BCSCs) [18], pancreatic cancer stem cells (PCSCs) [19], chronic myeloid leukemia (CML) [20] and acute myeloid leukemia (AML) stem cells [21] and acted as an important promotors in the maintenance and self-renew properties of these CSCs.
High expression of SIRT1 was found in CD133-positive glioma cells, which played an important role in tumor recovery and radiotherapy resistance. Knockdown of SIRT1 expression in these cells enhanced radiosensitivity and radiation-induced apoptosis in vitro and the mean survival rate of radiotherapy-treated mice bearing glioblastoma-CD133-positive tumors was significantly improved by SIRT1 knockdown as well [22]. Further study showed that SIRT1 increased expression in GSCs (CD133, sex determining region Y-box 2 (Sox2), and nestin positive) and cancerous neural stem cells (CNSCs) and played an important role in escaping p53-dependent tumor surveillance, thus induced oncogenic transformation and retained the neural cancer stemness of the cells [16]. In addition, SIRT1 expression was decreased in cells of glial type (glial fibrillary acidic protein (GFAP)-positive) after differentiation from GSCs [16]. Besides, another study also showed that translation of SIRT1 in nestin and notch intracellular domain (NICD)-positive GSCs was suppressed by cytoplasmic polyadenylation element binding protein 1 (CPEB1), a key modulator that induced differentiation at the post-transcriptional level [23]. These evidences indicated that SIRT1 played an essential role in GCSs and could be a therapeutic target of GSCs.
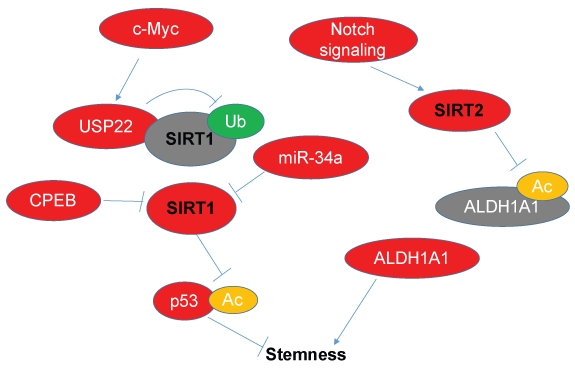
SIRT1 is overexpressed in human leukemia stem cells (LSC) and its inhibition increased apoptosis in LSC and suppressed proliferation of CML stem cells or primitive progenitor cells. Besides, inhibition SIRT1 in CML stem cells facilitated to eliminate CSCs by Imatinib, a breakpoint cluster region-c-Abl oncogene 1 (BCR-ABL) tyrosine kinase inhibitor (TKI), in part via p53 activation [20]. In SIRT1 homozygous knockout BALB/c mice models, CD150-negtive side population (SP) CML stem cells was also reduced and the maintenance of these cells was compromised. It seemed that SIRT1 knockout reduced cyclin-dependent kinase 6 (CDK6) expression and activated p53 through deacetylation instead of increasing its expression [24]. Except for CML stem cells, in primary human fms-like tyrosine kinase 3 internal tandem duplicated (FLT3-ITD) AML stem cells, SIRT1 was also selectively expressed in a high level and was enhanced by proto-oncogene c-MYC-induced high expression of ubiquitin specific peptidase 22 (USP22), a deubiquitinase (Figure 2). Besides, SIRT1 inhibition combined with FLT3 inhibitors promoted reduction of FLT3-ITD AML stem cells [21]. These evidences indicated that inhibition of SIRT1 network combined with TKIs or FLT3 inhibitors could be an attractive approach for BCR-ABL CML and FLT3-ITD AML treatment.
.
Figure 2: Schematic overview of molecular mechanisms of SIRT1/2 in CSCs. Schematic overview of molecular mechanisms of SIRT1/2 in CSCs, Ub: ubiquitination (green), Ac: acetylation (yellow), activated molecule (red), inactiviated molecule (grey).
View Figure 2
In addition, low level of miR-34a and high level of its target SIRT1 were found in CD44 positive and CD24-negtive BCSCs, and further study showed that miR-34a inhibited proliferative potential of BCSCs and downregulated the expression of CSC markers aldehyde dehydrogenase 1 (ALDH1), polycomb ring finger proto-oncogene Bmi1 and homeobox transcription factor Nanog partially by downregulating SIRT1 (Figure 2) [18]. Such mechanism might also occur in PCSCs [19]. In addition, many colorectal cancer specimens had strong SIRT1 expression, which had an obvious correlation with poor prognosis of colorectal cancer patients. Besides, SIRT1 was highly expressed in colorectal CSC-like cells and its expression had a co-localization with CD133, a current common used marker of CCSCs. Further study showed that SIRT1 knockdown reduced proportion of CD133-positive cells, dampened colony and sphere formation in vitro, and restrained tumorigenicity in vivo. Importantly, the expressions of Oct4, Cripto, Nanog, telomerase reverse transcriptase (TERT) and Lin28, which are stemness-associated genes, were decreased by SIRT1 deficiency [17]. These evidence indicated that SIRT1 played a crucial role in keeping the characteristics of stemness in colorectal cancer cells and SIRT1 may act as a potential prognostic factor of tumor recovery risk for cancer patients, and will contribute to providing a new therapeutic method for cancer treatment.
SIRT2 was another deacetylase that is increasingly identified as a essential regulator of cell cycle, cellular necrosis and cytoskeleton reorganization [25]. It’s recently reported to play an important role in carcinogenesis [26] and early lineage commitment of embryonic stem cell [27]. Recent research showed that SIRT2 could be induced by Notch signaling, thus leading to aldehyde dehydrogenase 1A1 (ALDH1A1) deacetylation and enzymatic activation to promote BCSCs (Figure 2) [28]. SIRT2 was also highly expressed in CSCs and its activity is required for CSCs proliferation arrest induced by resveratrol, a potent SIRT1 inhibitor [29]. However, in skin tumors but not normal skin, SIRT2 deletion up-regulates the stem cell marker CD34 [30]. No doubt, much more exploration should be conducted to make sure the exactly role of SIRT2 in CSCs.
Other members of sirtuins also have been demonstrated to play an important role in tumorigenesis and cancer development [31]. For example, SIRT3 limits reactive oxygen species (ROS) levels in cancer cell, thus induces the degradation of hypoxia inducible factor 1α (HIF-1α) and high level of SIRT3 may suppress intracellular acidification, inhibit Bcl2-associated X protein (BAX) activation and subsequent mitochondrial permeability transition (MPT) induction [32]. SIRT4 inhibits mitochondrial glutamine metabolism and responses to deoxyribonucleic acid (DNA) damage in cancer cells [33]. SIRT6 regulates glucose metabolism in tumors and acts as a tumor suppressor [34]. SIRT7 deacetylates H3K18 and maintains oncogenic transformation [35]. In addition, these sirtuins, especially SIRT6 [36,37], SIRT7 [38] and SIRT3 [39], also have been demonstrated to play important roles in the maintenance and differentiation of normal stem cells. They might also act as key regulators in CSCs, but their roles are remained to be further explored.
CSCs are a small subset of cells that are responsible for initiation, development, and recurrence of tumors. Therefore, it is important to understand the molecular mechanism of CSCs for translational applications using CSCs in the treatment of patients with cancer. Sirtuins are an important family of HDACs and have been shown to tightly relate to CSCs and play essential roles in the maintenance and self-renew properties of CSCs. These evidences indicated that sirtuins must be important pharmaceutical targets for cancer therapy.
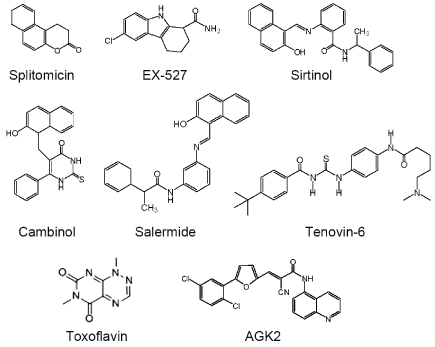
During past several years, some sirtuins inhibitors were indentified and developed to anti-cancer drug candidates, including splitomicin analogs, the indole derivative EX-527 (selisistat), sirtinol, cambinol, salermide, tenovins, toxoflavin, and AGK2 [40] (Figure 3). Several splitomicin derivatives were shown to inhibit SIRT2 and exerted antiproliferative properties in MCF7 breast cancer cells [41]. Another small molecular compound, EX-527 is a cell-permeable, specific inhibitor of SIRT1 and shown to increase acetylation at Lys382 of p53 and activate it [42]. Above all, a potent human SIRT1/2 inhibitor, sirtinol [43] was shown to induce senescence-like growth arrest in breast cancer cells, lung cancer cells [44], and enhance chemosensitivity to cisplatin and camptothecin in prostate cancer cells [45]. Besides, Cambinol and salermide are also SIRT1/2 inhibitors and showed anticancer properties [46,47]. Recently, some studies also explored salermide derivatives as sirtuins inhibitors and tested its efficient in cancer stem cells [48,49]. Tenovin-6 was identified as SIRT1/2/3 inhibitor [50] and antitumor was showed in acute lymphoblastic leukemia (ALL) [51], chronic lymphocytic leukemia (CLL) [52], gastric cancer [53] and colon cancer [54]. Besides, a SIRT1 specific inhibitor toxoflavin was also screened out and showed good anticancer effect [55]. Another SIRT2 potent and selective inhibitor AGK2 was also shown to induce apoptosis in glioma [56]. These studies indicated that sirtuins inhibitor may be efficient drug candidates for cancer therapy, but their effects on CSCs are remained to be explored.
In conclusion, sirtuins play essential roles in CSCs regulation and could be candidate molecular target in cancer treatment. However, the exactly function and mechanism of sirtuins have not been fully elucidated, much more work should be carried out.
References
-
Allegra A, Alonci A, Penna G, Innao V, Gerace D, et al. (2014) The cancer stem cell hypothesis: a guide to potential molecular targets. Cancer Invest 32: 470-495.
-
Meng X, Zhong J, Liu S, Murray M, Gonzalez-Angulo AM (2012) A new hypothesis for the cancer mechanism. Cancer Metastasis Rev 31: 247-268.
-
Francipane MG, Chandler J, Lagasse E (2013) Cancer Stem Cells: A Moving Target. Curr Pathobiol Rep 1: 111-118.
-
Visvader JE, Lindeman GJ (2008) Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer 8: 755-768.
-
Vincent A, Van Seuningen I (2012) On the epigenetic origin of cancer stem cells. Biochim Biophys Acta 1826: 83-88.
-
de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB (2003) Histone deacetylases (HDACs): characterization of the classical HDAC family. The Biochemical journal 370: 737-749.
-
Landry J, Slama JT, Sternglanz R (2000) Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem Biophys Res Commun 278: 685-690.
-
Tennen RI, Berber E, Chua KF (2010) Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mechanisms of ageing and development 131: 185-192.
-
Tennen RI, Bua DJ, Wright WE, Chua KF (2011) SIRT6 is required for maintenance of telomere position effect in human cells. Nat Commun 2: 433.
-
Rajendran R, Garva R, Krstic-Demonacos M, Demonacos C (2011) Sirtuins: molecular traffic lights in the crossroad of oxidative stress, chromatin remodeling, and transcription. J Biomed Biotechnol 368276.
-
Michishita E, Park JY, Burneskis JM, Barrett JC,Horikawa I (2005) Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Molecular biology of the cell 16: 4623-4635.
-
Finkel T, Deng CX, Mostoslavsky R (2009) Recent progress in the biology and physiology of sirtuins. Nature 460: 587-591.
-
Brooks CL, Gu W (2009) How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer 9: 123-128.
-
Ou X, Chae HD, Wang RH, Shelley WC, Cooper S, et al. (2011) SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation, and embryonic and adult hematopoiesis in the mouse. Blood 117: 440-450.
-
Deng CX (2009) SIRT, is it a tumor promoter or tumor suppressor? Int J Biol Sci 5: 147-152.
-
Lee JS, Park JR, Kwon OS, Lee TH, Nakano I, et al. (2015) SIRT1 is required for oncogenic transformation of neural stem cells and for the survival of "cancer cells with neural stemness" in a p53-dependent manner. Neuro-oncology 17: 95-106.
-
Chen X, Sun K, Jiao S, Cai N, Zhao X, et al. (2014) High levels of SIRT1 expression enhance tumorigenesis and associate with a poor prognosis of colorectal carcinoma patients. Sci Rep 4: 7481.
-
Ma W, Xiao GG, Mao J, Lu Y, Song B, et al. (2015) Dysregulation of the miR-34a-SIRT1 axis inhibits breast cancer stemness. Oncotarget 6: 10432-10444.
-
Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S (2011) Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PloS one 6: e24099.
-
Li L, Wang L, Li L, Wang Z, Ho Y, et al. (2012) Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell 21: 266-281.
-
Li L, Osdal T, Ho Y, Chun S, McDonald T, et al. (2014) SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute myeloid leukemia stem cells. Cell stem cell 15: 431-446.
-
Chang CJ, Hsu CC, Yung MC, Chen KY, Tzao C, et al. (2009) Enhanced radiosensitivity and radiation-induced apoptosis in glioma CD133-positive cells by knockdown of SirT1 expression. Biochemical and biophysical research communications 380: 236-242.
-
Yin J, Park G, Lee JE, Park JY, Kim TH, et al. (2014) CPEB1 modulates differentiation of glioma stem cells via downregulation of HES1 and SIRT1 expression. Oncotarget 5: 6756-6769.
-
Wang Z, Chen CC, Chen W (2015) CD150(-) Side Population Defines Leukemia Stem Cells in a BALB/c Mouse Model of CML and Is Depleted by Genetic Loss of SIRT1. Stem Cells 33: 3437-3451.
-
North BJ, Marshall BL, Borra MT, Denu JM, Verdin E (2003) The human Sir2 ortholog, SIRT, is an NAD+-dependent tubulin deacetylase. Mol Cell 11: 437-444.
-
Seo KS, Park JH, Heo JY, Jing K, Han J, et al. (2015) SIRT2 regulates tumour hypoxia response by promoting HIF-1alpha hydroxylation. Oncogene 34: 1354-1362.
-
Si X, Chen W, Guo X, Chen L, Wang G, et al. (2013) Activation of GSK3β by Sirt2 is required for early lineage commitment of mouse embryonic stem cell. PLoS One 8: e76699.
-
Zhao D, Mo Y, Li MT, Zou SW, Cheng ZL, et al. (2014) NOTCH-induced aldehyde dehydrogenase 1A1 deacetylation promotes breast cancer stem cells. The Journal of clinical investigation 124: 5453-5465.
-
Sayd S, Thirant C, El-Habr EA, Lipecka J, Dubois LG, et al. (2014) Sirtuin-2 activity is required for glioma stem cell proliferation arrest but not necrosis induced by resveratrol. Stem Cell Rev 10: 103-113.
-
Ming M, Qiang L, Zhao B, He YY (2014) Mammalian SIRT2 inhibits keratin 19 expression and is a tumor suppressor in skin. Exp Dermatol 23: 207-209.
-
Jeong SM, Haigis MC (2015) Sirtuins in Cancer: a Balancing Act between Genome Stability and Metabolism. Mol Cells 38: 750-758.
-
Chen Y, Fu LL, Wen X, Wang XY, Liu J, et al. (2014) Sirtuin-3 (SIRT3), a therapeutic target with oncogenic and tumor-suppressive function in cancer. Cell death & disease 5: e1047.
-
Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, et al. (2013) SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer cell 23: 450-463.
-
Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, Cosentino C, Greenson JK, MacDonald AI, McGlynn L, Maxwell F, Edwards J, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151: 1185-1199.
-
Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, et al. (2012) SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487: 114-118.
-
Etchegaray JP, Chavez L, Huang Y, Ross KN, Choi J, et al. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nature cell biology 17: 545-557.
-
Pan H, Guan D, Liu X, Li J, Wang L, et al. (2016) SIRT6 safeguards human mesenchymal stem cells from oxidative stress by co-activating NRF2. Cell Res.
-
Mohrin M, Shin J, Liu Y, Brown K, Luo H, et al. (2015) Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 347: 1374-1377.
-
Wang XQ, Shao Y, Ma CY, Chen W, Sun L, et al. (2014) Decreased SIRT3 in aged human mesenchymal stromal/stem cells increases cellular susceptibility to oxidative stress. J Cell Mol Med 18: 2298-2310.
-
Kozako T, Suzuki T, Yoshimitsu M, Arima N, Honda S, et al. (2014) Anticancer agents targeted to sirtuins. Molecules (Basel, Switzerland) 19: 20295-20313.
-
Neugebauer RC, Uchiechowska U, Meier R, Hruby H, Valkov V, et al. (2008) Structure-activity studies on splitomicin derivatives as sirtuin inhibitors and computational prediction of binding mode. Journal of medicinal chemistry 51: 1203-1213.
-
Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, et al. (2006) Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Molecular and cellular biology 26: 28-38.
-
Grozinger CM, Chao ED, Blackwell HE, Moazed D, Schreiber SL (2001) Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. The Journal of biological chemistry 276: 38837-38843.
-
Ota H, Tokunaga E, Chang K, Hikasa M, Iijima K, et al. (2006) Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene 25: 176-185.
-
Kojima K, Ohhashi R, Fujita Y, Hamada N, Akao Y, et al. (2008) A role for SIRT1 in cell growth and chemoresistance in prostate cancer PC3 and DU145 cells. Biochemical and biophysical research communications 373: 423-428.
-
Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, et al. (2006) Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer research 66: 4368-4377.
-
Lara E, Mai A, Calvanese V, Altucci L, Lopez-Nieva P, et al. Salermide, a Sirtuin inhibitor with a strong cancer-specific proapoptotic effect. Oncogene 28: 781-791.
-
Rotili D, Tarantino D, Nebbioso A, Paolini C, Huidobro C, et al. (2012) Discovery of salermide-related sirtuin inhibitors: binding mode studies and antiproliferative effects in cancer cells including cancer stem cells. Journal of medicinal chemistry 55: 10937-10947.
-
Rotili D, Tarantino D, Carafa V, Paolini C, Schemies J, Jung M, et al. (2012) Benzodeazaoxaflavins as sirtuin inhibitors with antiproliferative properties in cancer stem cells. Journal of medicinal chemistry 55: 8193-8197.
-
Lain S, Hollick JJ, Campbell J, Staples OD, Higgins M, et al. (2008) Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell 13: 454-463.
-
Jin Y, Cao Q, Chen C, Du X (2015) Tenovin-6-mediated inhibition of SIRT1/2 induces apoptosis in acute lymphoblastic leukemia (ALL) cells and eliminates ALL stem/progenitor cells. BMC Cancer 15: 226.
-
MacCallum SF, Groves MJ, James J, Murray K, Appleyard V, et al. (2013) Dysregulation of autophagy in chronic lymphocytic leukemia with the small-molecule Sirtuin inhibitor Tenovin-6. Scientific reports 3: 1275.
-
Hirai S, Endo S, Saito R, Hirose M, Ueno T, et al. (2014) Antitumor effects of a sirtuin inhibitor, tenovin-6, against gastric cancer cells via death receptor 5 up-regulation. PloS one 9: e102831.
-
Ueno T, Endo S, Saito R, Hirose M, Hirai S, Suzuki H, Yamato K and Hyodo I. The sirtuin inhibitor tenovin-6 upregulates death receptor 5 and enhances cytotoxic effects of 5-fluorouracil and oxaliplatin in colon cancer cells. Oncology research 21: 155-164.
-
Choi G, Lee J, Ji JY, Woo J, Kang NS, et al. (2013) Discovery of a potent small molecule SIRT1/2 inhibitor with anticancer effects. Int J Oncol 43: 1205-1211.
-
He X, Nie H, Hong Y, Sheng C, Xia W, et al. (2012) SIRT2 activity is required for the survival of C6 glioma cells. Biochem Biophys Res Commun 417: 468-472.