Journal of Genetics and Genome Research
Microevolutionary History of Helicobacter pylori During Infection: A Review
Mendoza-Elizalde S1,2, Olivares-Cervantes AL1, Zuñiga G2*, Valencia-Mayoral P3, Vigueras-Galindo JC1 and Velázquez-Guadarrama N1*
1Laboratorio de Infectología, Hospital Infantil de México Federico Gómez, México
2Laboratorio de Variación Biológica y Evolución, Departamento de Zoología, Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, México
3Dirección de Planeación, Hospital Infantil de México Federico Gómez, México
*Corresponding authors:
Norma Velázquez-Guadarrama, Laboratorio de Infectología, Hospital Infantil de México Federico Gómez, Dr. Márquez 162, Col. Doctores, Cuauhtémoc, México D.F., 06720, México, Tel: +52(55) 52289917x2081, Fax: +52(55) 55884549, E-mail: normave@himfg.edu.mx;
Gerardo Zuñiga, Departamento de Zoología, Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, Prol. Carpio esq. Plan de Ayala s/n, Col. Sto. Tomás, Mexico D.F., 11340, Mexico, Tel: +52(55) 57296300x62418, E-mail: capotezu@hotmail.com
J Genet Genome Res, JGGR-3-022, (Volume 3, Issue 1), Review Article; ISSN: 2378-3648
Received: November 06, 2015 | Accepted: April 11, 2016 | Published: April 13, 2016
Citation: Mendoza-Elizalde S, Olivares-Cervantes AL, Zuñiga G, Valencia-Mayoral P, Vigueras-Galindo JC, et al. (2016) Microevolutionary History of Helicobacter pylori During Infection: A Review. J Genet Genome Res 3:022. 10.23937/2378-3648/1410022
Copyright: © 2016 Mendoza-Elizalde S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Helicobacter pylori is a bacterium that has evolved with humans, is transmitted from person to person and persistently colonizes the stomach. During its long coexistence with humans, H. pylori has developed complex strategies to limit the degree and extent of inflammation and damage to the gastric mucosa. It is capable of altering the physiology and immune response of the host, thereby allowing it to persist throughout life. Infection with H. pylori has been linked to such diseases as severe gastritis, peptic ulcers and gastric cancer. Differences in the incidence of gastric cancer across the world can be attributed to regionally different H. pylori genotypes, dietary factors, environmental factors, and host factors, as well as virulence gene polymorphisms (particularly cagA and vacA) in different strains. The colonization of an individual by more than one strain of H. pylori is common, and a recurrent infection during childhood provides substrates for the acquisition or recombination of new genetic sequences. This review explores the evolutionary history of H. pylori during infection recurrence and the genetic diversity in humans as well as in an animal model.
Keywords
H. pylori, Recurrence, Genetic diversity, Recombination
Introduction
Humans are the definitive host of H. pylori. Phylogeographic studies have indicated that humans have been colonized by H. pylori for ≥ 58,000 years, since the first migration out of Africa [1]. H. pylori is a Gram-negative, pleomorphic bacillus that is classified as a non-invasive bacterium because it does not cross the epithelial barrier. H. pylori is motile owing to the presence of four to six sheathed polar flagella. It grows in microaerophilic conditions and requires between 3 and 5 days of growth at 37°C for primary isolation; its colonies are translucent and 1 to 2 mm in diameter [2]. Its genome size averages 1.7 Mbp, has a G+C content of 35 to 40% and encodes 1500 proteins [3]. Approximately 40% of H. pylori isolates contain a 23.3 kb plasmid that contains no known virulence factors. Additionally, the bacterium possesses at least two operons for each ribosomal ribonucleic acid (rRNA): 16S, 23S and 5S [4].
Infection with H. pylori is acquired within the first years of life and is related to geographic location [5]. The course of the infection is highly variable. Most infected individuals remain asymptomatic; only 10-20% develops atrophic gastritis, and of these, less than 3% develop gastric cancer [6]. In developed countries, the prevalence of H. pylori infection varies from 1.2% to 12.2%; it is low in children and adolescents and higher in adults and the elderly. The infection rate is higher (80%) in developing countries [7]. The main factor that predisposes individuals to infection is socioeconomic status, with greater infection prevalence observed among individuals living in overcrowded or impoverished conditions [8].
In contrast to many other bacterial pathogens, H. pylori does not exhibit clonal propagation in different human populations or within groups of patients. An exception to this rule is transmission among families, where clonal infectious strains have been observed more frequently among mothers and children [9].
Children are particularly vulnerable to infection by H. pylori, which is transmitted by infected parents, especially mothers. However, little is known about how and when maternal transmission occurs in childhood, particularly whether it occurs before or after weaning [10].
The colonization of an individual by more than one strain of H. pylori is common, and a recurrent infection during childhood provides substrates for the acquisition or recombination of new genetic sequences. This review explores the microevolutionary history of H. pylori during infection recurrence and the genetic diversity in humans as well as in an animal model.
Recurrence of H. pylori Infection
Recurrence of H. pylori infection in children and adults has been proposed to have two mechanisms. 1) recrudescence: and reinfection. Recrudescence is defined as the apparent elimination of infection due to suppression but not successful eradication of the bacteria [11,12]. By contrast, reinfection occurs after successful eradication, when a patient is re-infected with a new strain or with the same strain at least one year after eradication (Figure 1) [13,14].
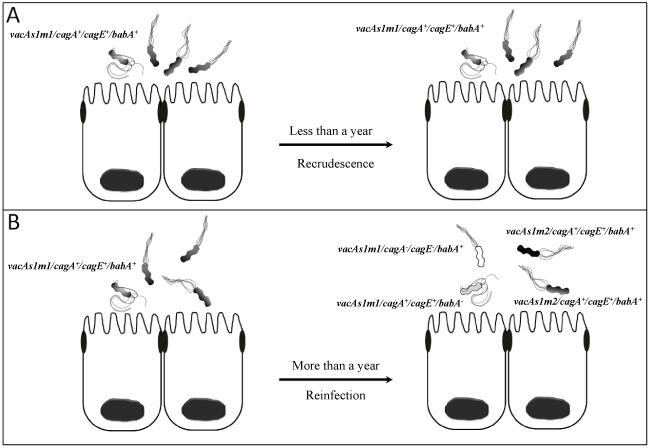
Figure 1: Recurrence of H. pylori infection in children and adults has been proposed to have two mechanisms. 1) Recrudescence: the apparent elimination of infection due to suppression but not successful eradication of the bacteria (infection with the same genotype strain by lack of compliance with treatment or antibiotic resistance), and 2) Reinfection:
occurs after successful eradication, when a patient is re-infected with a new strain or with the same strain at least one year after eradication (infection due to strains with different genotype and antimicrobial susceptibility).
View Figure 1
The effectiveness of antimicrobial treatment for H. pylori infection may be limited due to patient compliance with the treatment regimen, selective pressure for strains with high levels of resistance to antimicrobials of choice and high rates of recrudescence and reinfection. Furthermore, H. pylori infection recurrence is common in patients with low-efficiency treatments. Thus, treatment has been proposed to only temporarily suppress the bacteria and to be incapable of completely eradicating the infection from the host [14].
The annual recurrence rate of H. pylori is 2.67% in developed countries and 13% in developing countries such as Peru, Brazil, Chile, Vietnam and Bangladesh [11]. Many cases of recurrence in developed countries are due to late recrudescence within the first year (46%), while in developing countries the rate of reinfection is more pronounced and continues at the same pace as the eradication rate [13]. The rates of reinfection are extremely low in Europe and the USA; indeed, their rates are similar to that of South Korea, where the annual reported rate is approximately 1% [14].
Molecular biology techniques are very useful tools for the diagnosis of H. pylori infection. These techniques can identify the genotype of the bacteria before and after treatment and determine whether strains are genetically identical; thus, it has been confirmed that the recurrence of infection is due to recrudescence in 80% of cases [13]. However, the possibility of reinfection with an identical community strain that is shared among family members cannot be dismissed. Furthermore, different strains may be isolated from the same host, representing a broad diversification due to micro evolutionary changes in the host itself. This microevolution is driven by changes in the genetic background of a bacterial species that results in changes in various genes over time and can result in the presence of several existing genotypes in a single H. pylori population.
Genetic Diversity of H. pylori
Genetic diversity is the sum of the genetic information that exists among the members of a species. H. pylori is a bacterium with high genetic diversity. Typically, high genetic diversity in bacteria is attributed to the ability to easily incorporate exogenous DNA, to the possession of genes with a high mutation rate, to the ability to horizontally transfer genes and to recombination events [6,15]. Several studies have suggested that the acquisition and loss of genes are mechanisms that facilitate the adaptation of the bacterium to new host environments beyond the changes that occur in the gastric environment [16-23]. Likewise, specificity, inflammatory responses, and the availability and distribution of adhesion molecules in the human host are factors that may select for divergence between strains [24].
Possible scenarios have been proposed to account for the generation of variability in H. pylori: monoclonal and polyclonal transmission. In the first scenario, a single clone may diverge rapidly within the microniches of the stomach to adapt to the new environment; however, as soon as the niches are occupied, the genetic exchange rate is reduced. Thus, genetic changes can occur in a stepwise manner when there is variation in selection pressures. According to this scenario, most of the genetic variability of the clone was generated during the infancy of the patient (assuming that he or she was infected early in life), and therefore the clone was present at the initial medical examination [9]. In polyclonal transmission, clonal variants are gradually developed over several generations in the host as a divergent population; consequently, the genetic variability of the clones observed in any host is the sum of all of the accumulated variations in all transmissions from previous infections [9].
Recombination events between genes from the same strain or between different alleles in the genome of H. pylori have been suggested to contribute to the high diversity of strains in the host. Therefore, sub clones of the bacterium may differ both in genotype and phenotype despite having been isolated from the same biopsy [9]. Israel et al. (2001) [25] demonstrated that all the sub clones in an individual are unique within the biopsies and between biopsies from different parts of the stomach. Mendoza-Elizalde et al. (2015) [26] found that 27 of 32 Mexican paediatric patients presented sub clones of H. pylori with different genotypes, thereby confirming the extensive allelic diversity of the bacteria within a single patient.
Diversity between Nucleotide Sequences of H. pylori
The diversity of H. pylori nucleotide sequences exceeds that of other studied bacteria. The most unusual feature of this diversity is the high number of unique nucleotide sequences for each gene studied. Kansau et al. (1996) [27] analysed the nucleotide sequences of the glmM gene (encoding glutamate racemase) in 29 strains of H. pylori and found only two strains with identical glmM sequences.
The DNA sequences of virulence and housekeeping genes in H. pylori have different genotypes that may predominate in different human populations. Global studies have shown that high degrees of polymorphism in housekeeping genes is associated with changes in the third nucleotide position of codons; therefore, much of the observed variation is synonymous [28-30]. Mendoza-Elizalde (2015b) [31] observed a high degree of polymorphism in the housekeeping genes of 80 H. pylori strains from two Mexican paediatric patients, revealing 26 new alleles for the atpA, efp, mutY, ppa, trpC, ureI and yphC genes.
Phylogenetic analysis based on housekeeping gene sequences provides an overview of the evolutionary dynamics of infectious H. pylori strains within individual families [32]. These studies have revealed the presence of five ancestral populations and subpopulations of H. pylori: H. pylori Europe (hpEurope), H. pylori Asia 2 (hpAsia2), H. pylori North East Africa (hpNEAfrica), H. pylori Africa 1 (hpAfrica1) and H. pylori East Asia (hpEAsia) (Figure 2) [1,33-35].
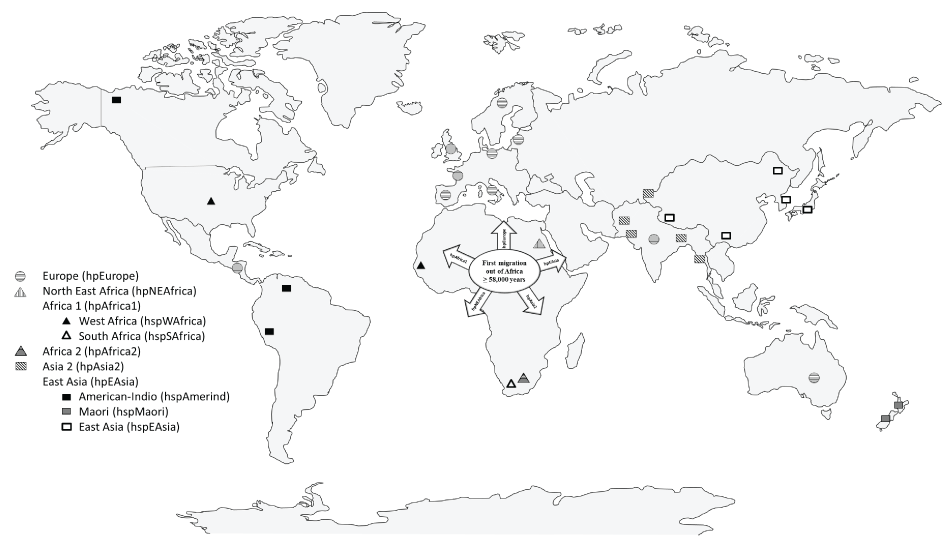
Figure 2: Ancestral populations and subpopulations of Helicobacter pylori . H. pylori Europe (hpEurope), H. pylori Asia 2 (hpAsia2), H. pylori North East Africa (hpNEAfrica), H. pylori Africa 1 (hpAfrica1) with the subpopulations H. pylori West Africa (hspWAfrica) and H. pylori South Africa (hspSAfrica), H. pylori Africa 2 (hpAfrica2), and H. pylori East Asia (hpEAsia), with the subpopulations H. pylori American-Indio (hspAmerind), H. pylori Maori (hspMaori), and H. pylori East Asia (hspEAsia).
View Figure 2
Bacterial Strain-Host Interactions
Humans can be colonized by a single strain or multiple strains of H. pylori. Over time, the evolutionary dynamics of the organism itself can give rise to multiple subpopulations of H. pylori. Processes such as mutation or genetic recombination play a fundamental role in this diversity [30], thereby allowing the emergence of different genotypes such as genotype transitions (presence of chimaeras), virulent or toxigenic genotypes and non-virulent or non-toxigenic genotypes.
It has been observed that recombination of H. pylori over long periods of time may give rise to new clones. For example, strains with the cagA gene, which belongs to the pathogenicity island (cagPAI+), can pass cagA+ to cagA- strains through the acquisition of genetic material from cagA- strains (cagPAI-) [36,37]. These arrangements have not been well studied and are detectable only during the transition phase, after passage of an in vitro strain (culture) to an in vivo environment (animal) or during colonization of an uninfected host [36].
A model has been proposed to explain the evolution and transmission of H. pylori. This model assumes that strains containing the virulence genes encoding the oncogenic protein CagA, the vacuolating cytotoxin VacA, and the adhesin BabA represent a "state of maximum welfare" and that the activation of any of these genes reduces the physical state of the bacterium. H. pylori lives in the stomach of an individual for many years, where it continually produces strains that are cag, vacA or babA positive or -negative [38]. The emergence of multiple defective derivatives in some cases may overcome the wild-type bacterium, allowing the isolation of cag+ and cag- strains from the same patient. However, these defective derivatives do not survive for long times and thus represent the loss of some branches of the evolutionary tree. Therefore, only strains that are efficient in long-term colonization and can participate in person-to-person transmission will endure and direct the evolution of the species [37,38].
Animal Model of H. pylori Infection
As mentioned above, H. pylori displays a high level of genetic diversity due to high rates of mutation and recombination that allow the generation of variability between different strains. The use of animal models that are informative and manageable allows control over the presence or absence of bacterial antigens during different stages of infection. Therefore, animal models are routinely used to study the relationships between the immune response or basic genetics and susceptibility to bacterial infection and to identify factors related to survival and growth [23,39].
Microbial populations in infected or colonized hosts are subjected to selective forces and therefore are dynamic. The population dynamics of H. pylori during colonization of an infected animal offers an experimental model in which phenotypic and genotypic evolution are quantifiable in vivo, from the initial inoculation until the definitive establishment of a quasi species [40]. The spectrum of organisms used to study the pathogenesis of H. pylori includes rhesus monkeys, mice, and gerbils [41,42]. Since Hirayama et al. (1996) [43] reported the establishment of a persistent H. pylori infection in Mongolian gerbils (Meriones unguiculatus), research has been conducted on the association between H. pylori and gastric diseases. Some findings have suggested that H. pylori is directly related to gastric carcinogenesis [42].
The Mongolian gerbil infection model is considered useful because the changes observed in the gastric mucosa in this model are similar to those seen in human stomachs after infection with H. pylori [44]. Nakagawa et al. (2005) [42] reported that gastric colonization by H. pylori in Mongolian gerbils is related to several bacterial factors, including urease production and motility. Iwao et al. (1999) [45] reported that H. pylori motility was an important factor for the gastric colonization of Mongolian gerbils and that there were differences in gastric colonization due to phenotypic differences between the strains in the gerbils. Karita et al. (1991) [46] and Ohkusa et al. (2003) [47] observed that different strains induced different levels of infection severity in Mongolian gerbils. Furthermore, the cag PAI has been reported to play an essential role in the inflammatory process and formation of ulcers in vivo in Mongolian gerbils [48].
The microevolutionary history of H. pylori infection in humans reveals a remarkably high level of genetic diversity in this bacterium that is mainly generated by point mutations and by recombination (intra genomic or inter genomic) [49,50]. In turn, this high level of diversity has allowed the identification of different strains, sub clones or genotypes in the same individual that may occupy different microniches in the stomach. These strains may have selective advantages at different times during colonization and may allow the emergence of new bacterial populations [51]. Additionally, the use of an animal model has allowed the analysis of microevolutionary changes in the bacterium that occur during colonization, persistence and induction of disease in animals.
Acknowledgements
The authors gratefully acknowledge the Hospital Infantil de México Federico Gómez, Consejo Nacional de Ciencia y Tecnología (CONACyT), Posgrado de Ciencias Quimicobiológicas (ENCB-IPN), Secretaria de Investigación y Posgrado (SIP). This work was part of MESs PhD dissertation and the first author was a CONACyt (216177) fellow. Federal Resources (HIM/2011/ 080 SSa. 1005) from SSA supported this project.
References
-
Linz B, Balloux F, Moodley Y, Manica A, Liu, et al. (2007) An African origin for the intimate association between humans and Helicobacter pylori. Nature 445: 915-918.
-
Algood HM, Cover TL (2006) Helicobacter pylori persistence: an overview of interactions between H. pylori and host immune defenses. Clin Microbiol Rev 19: 597-613.
-
Suerbaum S, Mitchetti P (2002) Helicobacter pylori Infection. N Engl J Med 347: 1175-1186.
-
Kusters JG, AHM van Vliet, EJ Kuipers (2006) Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev 19: 449-490.
-
Cover TL, Blaser MJ (2009) Helicobacter pylori in health and disease. Gastroenterology 136: 1863-1873.
-
Costa AC, Figueiredo C, Touati E (2009) Pathogenesis of Helicobacter pylori infection. Helicobacter 14: 15-20.
-
Rajindrajith S, Devanarayana NM, Janaka de Silva H (2009) Helicobacter pylori infection in children. Saudi J Gastroenterol 15: 86-94.
-
Weyermann M, Rothenbacher D, Brenner H (2009) Acquisition of Helicobacter pylori infection in early childhood: independent contributions of infected mothers, fathers and siblings. Am J Gastroenterol 104: 182-189.
-
Lundin A, Björkholm B, Kupershmidt I, Unemo M, Nilsson P, et al. (2005) Slow Genetic Divergence of Helicobacter pylori Strains during Long-Term Colonization. Infect Immun 73: 4818-4822.
-
Tanih NF, Ndip LM, Ndip RN (2011) DNA sequence analysis of South African Helicobacter pylori Vacuolating Cytotoxin Gene (vacA). Int J Mol Sci 12: 7459-7468.
-
Soto G, Bautista CT, Roth DE, Gilman RH, Velapatiño B, et al. (2003) Helicobacter pylori reinfection is common in Peruvian adults after antibiotic eradication therapy. J Infect Dis 188: 1263-1275.
-
McMahon BJ, Bruce MG, Hennessy TW, Bruden DL, Sacco F, et al (2006) Reinfection after successful eradication of Helicobacter pylori: a 2-year prospective study in Alaska Natives. Aliment Pharmacol Ther 23: 1215-1223.
-
Niv Y ( 2008) Helicobacter pylori recurrence after successful eradication. 14: 1477-1488.
-
Ryu KH, Yi SY, Na YJ, Baik SJ, Yoon SJ, et al. (2010) Reinfection rate and endoscopic changes after successful eradication of Helicobacter pylori. World J Gastroenterol 16: 251-255.
-
Hussain MA, Kauser F, Khan AA, Tiwari S, Habibullah CM, et al. (2004) Implications of molecular genotyping of Helicobacter pylori isolates from different human populations by genomic fingerprinting of enterobacterial repetitive intergenic consensus regions for strain identification and geographic evolution. J Clin Microbiol 42: 2372-2378.
-
Suerbaum S (2000) Genetic variability within Helicobacter pylori. Int J Med Microbiol 290: 175-181.
-
Blaser MJ, Berg DE (2001) Helicobacter pylori genetic diversity and risk of human disease. J Clin Invest 107: 767-773.
-
Blaser MJ, Atherton JC (2004) Helicobacter pylori persistence: biology and disease. J Clin Invest 113: 321-333.
-
Raymond J, Thiberg JM, Chevalier C, Kalach N, Bergeret M, et al. (2004) Genetic and transmission analysis of Helicobacter pylori strains within a family. Emerg Infect Dis 10: 1816-1821.
-
Giono-Cerezo S, Camorlinga PM, Aguilar GR (2006) Diagnóstico microbiológico, serológico, genotipificación de Helicobacter pylori aislado de biopsias de niños y adultos. Detección molecular de la isla de patogenicidad cag de Helicobacter pylori. Rev Latinoam Microbiol 48: 99-104.
-
Kivi M, Rodin S, Kupershmidt I, Lundin A, Tindberg Y, et al. (2007) Helicobacter pylori genome variability in a framework of familial transmission. BMC Microbiol 7: 1-8.
-
Salama NR, Gonzales-Valencia G, Deatherage B, Aviles-Jimenez F, Atherton JC, et al. (2007) Genetic analysis of Helicobacter pylori strain populations colonizing the stomach at different times postinfection. J Bacteriol 189: 3834-3845.
-
Peek RM (2008) Helicobacter pylori infection and disease: from humans to animal models. Dis Model Mech 1: 50-55.
-
Gangwer KA, Shaffer CL, Suerbaum S, Lacy DB, Cover TL et al. (2010) Molecular evolution of the Helicobacter pylori vacuolating toxin gene vacA. J Bacteriol 192: 6126-6135.
-
Israel DA, Salama N, Krishna U, Rieger UM, Atherton JC, et al. (2001) Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc Natl Acad Sci USA 98: 14625-14630.
-
Mendoza-Elizalde S, Cortes-Marquez AC, Giono-Cerezo S, Zuniga G, Consuelo-Sanchez A, et al. (2015) Analysis of the genotypic diversity of strains of Helicobacter pylori isolated from pediatric patients in Mexico. Infect Genet Evol 29: 68-74.
-
. Kansau I, Raymond J, Bingen E, Courcoux P, Kalach N, et al. (1996) Genotyping of Helicobacter pylori isolates by sequencing of PCR products and comparison with the RAPD technique. Res Microbiol 147: 661-669.
-
Achtman M, Azuma T, Berg DE, Ito Y, Morelli G, et al. (1999) Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol 32: 459-470.
-
Morelli G, Didelot X, Kusecek B, Schwarz S, Bahlawane C, et al. (2010) Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genet 6: e1001036.
-
Secka O, Antonio M, Berg DE, Tapgun M, Bottomley C, et al. (2011) Mixed infection with cagA positive and cagA negative strains of Helicobacter pylori lowers disease burden in The Gambia. PLoS One 6: e27954.
-
Mendoza-Elizalde S. Microevolución de cepas de Helicobacter pylori aisladas de pacientes pediátricos (Tesis Doctoral). Instituto Politécnico Nacional-Escuela Nacional de Ciencias Biológicas, México 2015b; 1-151.
-
Raymond J, Thiberge JM, Kalach N, Bergeret M, Dupont C, et al. (2008) Using macro-arrays to study routes of infection of Helicobacter pylori in three families. PLoS One 3: e2259.
-
Atherton JC, Sharp PM, Cover TL, Gonzalez-Valencia G, Peek RM Jr, et al. (1999) Vacuolating cytotoxin (vacA) alleles of Helicobacter pylori comprise two geographically widespread types, m1 and m2, and have evolved through limited recombination. Curr Microbiol 39: 211-218.
-
Atherton JC and Blaser MJ (2009) Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J Clin Invest 119: 2475-2487.
-
Latifi-Navid S, Ghorashi SA, Siavoshi F, Linz B, Massarrat S, et al. (2010) Ethnic and Geographic Differentiation of Helicobacter pylori within Iran. PLoS One 5: e9645.
-
Ferrero RL, Jenks PJ, (2001) In vivo adaptation to the host..In: Mobley HLT, Mendz GL, Hazell SL, Helicobacter pylori: physiology and genetics. ASM , Washington, D.C. 583-592.
-
Prouzet-Mauleon V, Abid MH, Lamouliatte H, Kauser F, Megraud F, et al. (2005) Pathogen evolution in vivo: genome dynamics of two isolates obtained 9 years apart from a duodenal ulcer patient infected with a single Helicobacter pylori strain. J Clin Microbiol 43: 4237-4241.
-
Montecucco C, Rappuoli R (2001) Living dangerously: How Helicobacter pylori survives in the human stomach. Nat Rev Mol Cell Biol 2: 457-466.
-
O'Rourke JL, Lee A (2003) Animal models of Helicobacter pylori infection and disease. Microbes Infect 5: 741-748.
-
Webb GF, Blaser MJ (2002) Dynamics of bacterial phenotype selection in a colonized host. Proc Natl Acad Sci USA 99: 3135-3140.
-
Boonjakuakul JK, Canfield DR, Solnick JV (2005) Comparasion of Helicobacter pylori virulence gene expression in vitro and in the Rhesus macaque. Infect Immun 73: 4895-4904.
-
Nakagawa S, Osaki T, Fujioka Y, Yamaguchi H, Kamiya S (2005) Long-term infection of mongolian gerbils with Helicobacter pylori: microbiological, histopathological, and serological analyses. Clin Diagn Lab Immunol 12: 347-353.
-
Hirayama F, Takagi S, Yokoyama Y, Iwao E, Ikeda Y (1996) Establishment of gastric Helicobacter pylori infection in Mongolian gerbils. J Gastroenterol 9: 24-28.
-
Tatematsu M, Nozaki K, Tsukamoto T (2003) Helicobacter pylori infection and gastric carcinogenesis in animal models. Gastric Cancer 6: 1-7.
-
Iwao E, Hirayama F, Takagi S, Yokoyama Y, Ikeda Y (1999) Virulence factors of Helicobacter pylori affecting its gastric colonization in Mongolian gerbils. J Gastroenterol 34: 47-54.
-
Karita M, Kouchiyama T, Okita K, Nakazawa T (1991) New small animal model for human gastric Helicobacter pylori infection: success in both nude and euthymic mice. Am J Gastroenterol 86: 1596-1603.
-
Ohkusa T, Okayasu I, Miwa H, Ohtaka K, Endo S, et al. (2003) Helicobacter pylori infection induces duodenitis and superficial duodenal ulcer in Mongolian gerbils. Gut 52: 797-803.
-
Akanuma M, Maeda S, Ogura K, Mitsumo Y, Hirata Y, et al. (2002) The evaluation of putative virulence factors of Helicobacter pylori for gastroduodenal disease by use of a short-term Mongolian gerbil infection model. J Infect Dis 185: 341-347.
-
Suerbaum S, Smith JM, Bapumia K, Morelli G, Smith NH, et al. (1998) Free recombination within Helicobacter pylori. Proc Natl Acad Sci. USA 95: 12619-12624.
-
Falush D, Kraft C, Taylor NS, Correa P, Fox JG, et al. (2001) Recombination and mutation during longterm gastric colonization by Helicobacter pylori: estimates of clockrate, recombination size, and minimal age. Proc Natl Acad Sci USA 98: 15056-15061.
-
Suerbaum S and Josenhans C (2007) Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol 5: 441-452.





