International Journal of Cancer and Clinical Research
P53 Sensitizes Breast Cancer Stem Cells to Let-7 miRNAs Induced Repression
Jian Liu1, Ning Du1, Huangzhen Wang1, Xin Wang2, Peili Wang1, Jia Zhang1, Gang Li1, Jing Zhang1, Sida Qin1, Chongwen Xu1, Shou-Ching Tang3,4, Xin Sun1, Boxiang Zhang1, Dapeng Liu1 and Hong Ren1*
1Department of Thoracic Surgery and Oncology, The Second Department of Thoracic Surgery, The First Affiliated Hospital of Xi'an Jiaotong University, Xi'an, Shaanxi Province, China
2Department of Gastroenterology, The First Affiliated Hospital of Xi'an Jiaotong University, Xi'an, Shaanxi Province, China
3Georgia Regents University Cancer Center, 1411 Laney Walker Blvd, Augusta, Georgia, USA
4Tianjin Medical University Cancer Institute and Hospital, Tianjin, China
*Corresponding author:
Hong Ren, Department of Thoracic Surgery and Oncology, The Second Department of Thoracic Surgery, The First Affiliated Hospital of Xi'an Jiaotong University, 277 Yanta West Road, Xi'an, Shaanxi Province, 710061, P.R. China, Tel: +86-13909237351; E-mail: medicinedr.ren@gmail.com
Int J Cancer Clin Res, IJCCR-3-047, (Volume 3, Issue 2), Original Article; ISSN: 2378-3419
Received: August 26, 2014 | Accepted: March 07, 2016 | Published: March 10, 2016
Citation: Liu J, Du N, Wang H, Wang X, Wang P, et al. (2016) P53 Sensitizes Breast Cancer Stem Cells to Let-7 miRNAs Induced Repression. Int J Cancer Clin Res 3:047. 10.23937/2378-3419/3/2/1047
Copyright: © 2016 Liu J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: Cancer stem cells (CSCs) are responsible for the tumor recurrence and resistance to chemo-radiotherapy. Let-7 family of miRNAs functions in regulating cancer biology by repressing multiple downstream oncogenes.
Objective: In this study, we examined the regulatory loop of suppressive let-7 and p53 in the pathogenesis of breast cancer, and explored the mechanism through which this loop affects the growth patterns of breast cancer stem-like cells (BrCSCs).
Methods: The expressions of p53 and let-7 were detected by western blot and qRT-PCR. The tumor suppressive functions of p53 and let-7 were measured using cell viability and sphere formation assay.
Results: Both p53 and let-7 inhibited breast cancer cell growth and self-renewal of stem cells, and formed the let-7/p53 negative feedback loop. The inhibitory effects of p53 on cell growth are synergistic with that of let-7.
Conclusions: Both let-7 and p53 negatively regulate the growth of BrCSCs. In addition, their suppressive effects are additive. Our findings suggest that the existence of let-7/p53 negative feedback loop assists in regulating BrCSCs, and it may be used as a biomarker for the development of let-7 and p53, which is a targeted therapy in breast cancer. Furthermore, our data supports the dual targeting of let-7 and p53 in breast cancer development, and let-7 miRNAs could help to predict the prognosis of clinical chemotherapy response.
Keywords
Breast cancer; Cancer stem-like cells; Let-7 miRNAs; P53; Feedback loop; Therapy response
Introduction
Breast cancer is the most common and deadly malignancy among women worldwide, with the highest incidence in the United States and Europe [1]. The good news is that advanced medical science and technologies have given western countries effective strategies and methods of prevention and treatment of breast cancer, driving mortality rates steadily downward [1]. Breast cancer is one of the leading causes of cancer death among females in economically developing countries, and the mortality appears to be on the increase (China included) [2]. These countries didn’t share mortality decline [3]. MiRNAs based molecular therapy exhibited promising effects in anticancer treatments, for their roles in inducing cell death, regulating metastasis and drug resistance [4].
Let-7 family functioned as suppressive miRNAs, repressing malignancies through targeting oncogenic mRNAs, such as Ras, HMGA2, cyclin A/D, CDK4/6, c-Myc, LIN28, DICER1, NF-kB and so on [4]. They were demonstrated to exert effective repressions on tumor generation, proliferation, migration, relapse and therapy resistance, by binding to and degrading the targeted mRNAs through 3’UTR [4]. Let-7 miRNAs and their targeted genes could also form regulatory loops, such as let-7/Lin-28, let-7/IL-6, let-7/c-Myc, and with some newly defined ones. P53 is the key regulator in mediating cell death, cycle, and DNA damage response. P53 based chemotherapy has also been proven to be effective in anticancer treatment [5-7].
Tumors are groups of heterogeneous cells, containing some silent and resistant stem cells, made for tumor bulk survival through chemo-radiotherapy and to achieve a long-term tumor existence. Strategies specially targeting these CSCs bring hopes to sensitize cancer bulk and to anti-cancer treatments [8,9]. Many suppressive genes have been explored in let-7 regulated cancer stem cells abilities, and p53 was found to be related with stem cell functions by regulating multiple miRNAs [5,10]; but the interactions between p53 and let-7 in CSCs’ biology are still unclear. Previously, Lee et al. found that doxorubicin stimulated p53 could induce the let-7 expression through decreasing Lin28A [11]; whereas, another two groups identified that let-7 miRNAs were negatively correlated with p53 levels, which may directly bind to the enhancer in the promoter sequence of let-7 family members [12,13]. In our study, we explored the possible relationships between let-7 and p53, and studied their roles in regulating breast cancer and breast cancer stem-like cells (BrCSCs) respectively.
Materials and Methods
Cell culture and the infection of let-7 lentiviral
Human breast cancer cells MCF-7, ZR75-1, T47-D, MDA-MB-231, HS578T and NAFA were purchased from ATCC and kept at the Central Laboratory affiliated to the Medical College of Xi’an Jiaotong University, China. These cancer cells were kept and cultured in RPMI-1640 (cellgro, CORNING), and supplemented with 10% ~ 20% FBS, 1% penicillin, and streptomycin (Invitrogen). The suspending sphere were cultured in Ham’s F-12 and RPMI-1640 medium, supplemented with 20 μg/mL of epidermal growth factor (EGF), 20 ng/ml human basic fibroblast growth factor (HbFGF), 20 ng/mL of hydrocortisone, 6 μg/ml insulin, and 1% penicillin and streptomycin (Invitrogen) [14]. All cells were cultured in 5% CO2 at 37°C.
Oligonucleotides encoding let-7 miRNAs were synthesized and cloned into the pGLVU6/RFP vector (GenePharma Inc., Shanghai, China). The propagations of lentiviral vectors were conducted following the protocols applied in previous research [15,16]. Lentiviral plasmids and packaging vectors (PMD2/PSPAX2) were transfected into HEK 293T cells by Ca2+ phosphate transfection method. The viral supernatant was collected 48 hours post-transfection.
Cell viability and apoptosis rate detection
We explored the effects of let-7 miRNAs and p53 on cells by using MTT assay. Breast cancer cells of different groups were adjusted to 3~5 × 103 cells/well in the volume of 200 μl medium in 96-well plate first, and then incubated with different concentrations of Nutlin-3(N3, Sigma-Aldrich) for 24h, 48h, 72h and 96h respectively. Finally, these cells were cultured with 20 μl Methyl Thiazolyl Tetrazolium (MTT, 5 g/l) at 37°C for 4h. Each concentration had 5 repeats on the same plate. The optical density (OD) was detected, and the cell viability rates were calculated. Every sample was detected three times and presented as a mean value ± SEM based on independent experiments.
For apoptosis rates analysis, cancer cells of 5 × 104/ml in the logarithmic phase was collected and cultured in 6-well dishes at 1 × 105 cells/well. After treated with 0.05 mM doxorubicin or combined with N3 (the concentration was determined according to our previous results) [17], cells were then washed with PBS twice. Afterwards the cells were suspended in 500 μl of 1 × binding buffer, and then 5 μl Annexin V-FITC (BD Biosciences), 5 μl PI (20 μg/ml, BD Biosciences) were added. The mixture was gently vortexed and incubated for 15 min in the dark.
Sphere-formation assays
To observe the let-7 miRNAs and N3 functions, different groups were first treated with DMSO or N3 of different concentrations for 2 days, then were plated in 6cm or 10 cm ultra-low attachment dishes (Corning), in the presence of N3 [7]. On the 7th day, the spheres number and size were counted and measured under a low power field inverted microscope. The mammosphere formation efficiency (MFE) was calculated as the percentage ratio between obtained spheres and plated cells (mean ± SD of three independent experiments) [7]. Obtained spheres of different groups were disaggregated into single cells to acquire M2 cells, testing their ability of self-renewal and proliferation ability.
Detection of miRNAs and protein expression
The expression level of miRNA was examined using real-time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). MiR-U6 was taken as the control. The level of miRNA was calculated and presented following the 2-ΔΔCt method (ΔΔCt = ΔCt (positive)-ΔCt (control)). The membranes were incubated with specific primary antibodies after being blocked with (Santa Cruz); Abcam; Cell Signaling, p53 (FL393, sc-6243, sc-6243-G), Vinculin (4650) and β-Actin (4967) overnight; then secondary antibodies were conjugated with horseradish peroxidase (HRP) for mouse, rabbit and goat for 1 h (Santa Cruz). The western blot was scored as positive if the band of interest was present at the expected molecular weight. All analyses were performed in triplicate.
Statistical analysis
All statistical analyses were performed under the SPSS13.0 (IBM) and Excel (Microsoft). All data were represented as mean ± SD. Statistical analysis was conducted by using Student’s tests. The significance of each value was determined when P value was less than 0.05.
Results and Discussion
Let-7 inhibited the cell proliferation and self-renewal of stem-like cells
To explore the functions of let-7 on breast cancer stem cells, we first used lentiviral based let-7 miRNAs vectors (RFP) to construct multiple let-7 overexpressed breast cancer cells (Figure S1a). We first tested the proliferative inhibition of let-7 miRNAs by using MTT assay. The results showed that let-7a and let-7b exhibited the strongest suppressions among let-7 family of miRNA in ER positive breast cancer (Figure S1b), but with little inhibition on MDA-MB-231 cells (Figure S1b). We then enriched cancer stem cells of let-7 overexpressed cells through culturing spheres in ultra-low attachment plates. The results of mammosphere forming efficiency (MFE) of let-7 overexpressed MCF-7, ZR75-1 and T47-D cells were shown in figure 1a-b, and let-7a/b both inhibited the self-renewal of mammosphere of the 1st generation (compared to the control group, p < 0.01). Mammospheres from the 1st generation were disaggregated and suspended to form the 2nd generation of spheres. Results showed that let-7a/b inhibited the sphere size of both the 1st and 2nd generation (Figure S2a-b, compared to the control group, p < 0.05). The mean sizes of MCF-7, ZR75-1 and T47-D spheres were measured and calculated (red sphere meant that RFP based let-7 miRNAs were continually overexpressed, compared to the control group, p < 0.05, Figure S2c).
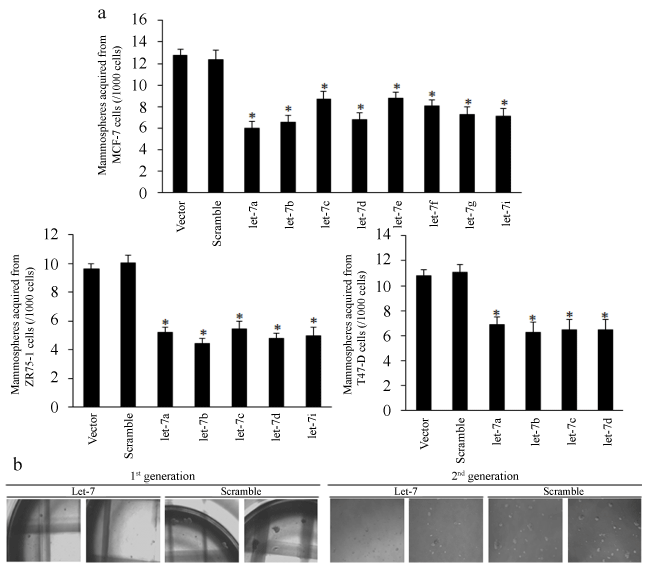
.
Figure 1: The repressive effects of let-7 on breast cancer stem-like cells. (a) Let-7a/b both inhibited the spheres number of the 1st generation effectively, p < 0.05, compared to the control group in MCF-7, ZR75-1 and T47-D cells; (b) The representative images of spheres of different generations.
View Figure 1
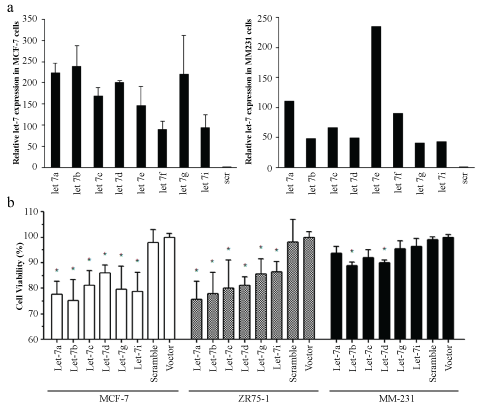
.
Figure S1: The suppressive effects of let-7 on breast cancer cells. (a) Let-7 expression levels were significantly higher than those of MCF-7and MDA-MB-231 Scramble and vector cells (p < 0.01); (b) The proliferation inhibitions of let-7 miRNAs on breast cancer cells (p < 0.05), and let-7aand let-7b exerted the most powerful inhibitions among let-7 family of miRNA. However, let-7 seemed to exert little inhibition on MDA-MB-231 cells.
View Figure S1
By western blot, we found that p53 expressions decreased unexpectedly in let-7 enforced cancer cells and cancer stem cells (Figure 2a). As p53 was identified as powerful tumor suppressor in cancer research, it’s indicated that let-7 miRNAs suppressed cancer malignancy is independent of tumor suppressive p53, which was previously identified to act as crucial tumor suppressors.
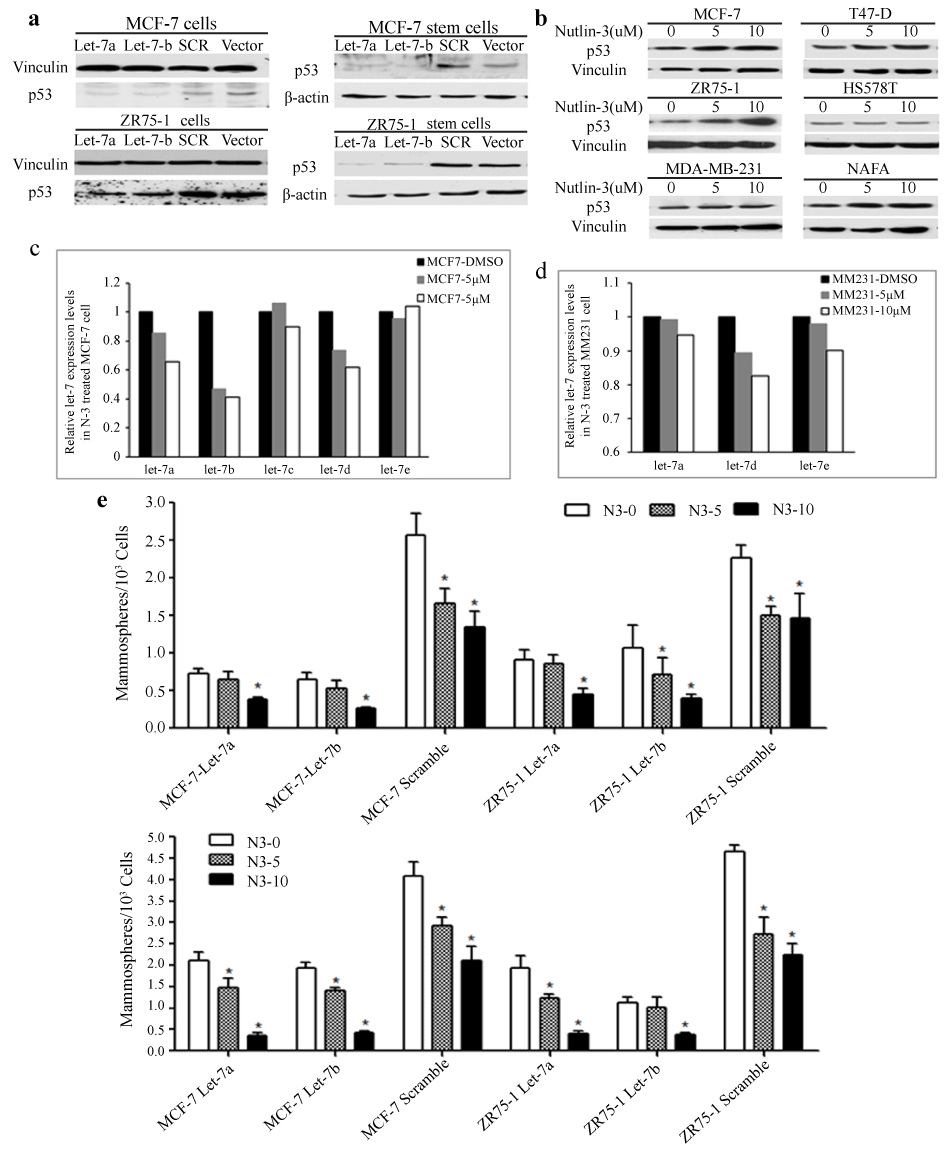
.
Figure 2: The effects of p53 and its combination with let-7 on spheres. (a) The results of western blot showed that p53 expressions decreased significantly in cancer cells and cancer stem cells depicting over expressed let-7 miRNAs; (b) The levels of expression of p53 in MCF-7, T47-D, ZR75-1, HS578T, MDA-MB-231 and NAFA cells; (c) Let-7 expressions in MCF-7 cells with increased wild type p53 levels, induced by different concentrations of Nutlin-3; (d) Let-7 expression levels in MDA-MB-231 cells; (e) The combination of let-7 and p53 showed strong inhibition on self-renewal of MCF-7 and ZR75-1 stem cells (above, 1st, below, 2nd).
View Figure 2
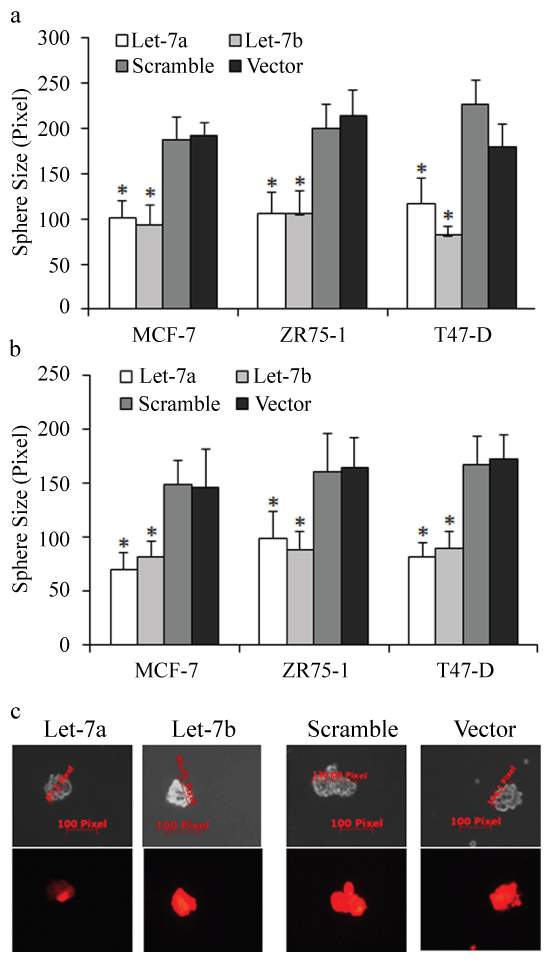
.
Figure S2: The effects of let-7 on spheres. (a-b) Let-7a/b inhibited the mammosphere sizes of the 1st and the 2nd generations of breast cancer stem-like cells; (c) The representative images of measurements of mammosphere size in different groups.
View Figure S2
The relationship between p53 and let-7 in breast cancer cells
We treated multiple kinds of breast cancer cells using different concentrations of Nutlin-3 to confirm the p53 expression alternations and levels. We found that p53 could be stimulated by N3 in MCF-7, T47-D, ZR75-1 and NAFA cell lines; however, in HS578T and MDA-MB-231 cell line, p53 expression didn’t change significantly with increased N3 concentration (Figure 2b). As wild type p53 only existed in MCF-7 and ZR75-1 cells, and let-7 expressions universally decreased in N3 treated MCF-7 (Figure 2c), but not MDA-MB-231 (Figure 2d) cells, meaning that lower expressions of let-7 miRNAs may act as indicators of p53 induced repression.
The inhibitory effects of p53 on breast cancer cells
To confirm the effects of p53 on breast cancer cells, we tested the inhibitory functions of p53 on cell proliferation by MTT assay. Increased p53 in MCF-7 and ZR75-1 cells both inhibited the proliferation significantly (compared to the control group, p < 0.05, Figure S3a). However, N3 also exerted inhibition on MDA-MB-231 cell growth, which may be independent of p53 status (Figure S3a).
Increased wild type p53 enhanced the inhibitory effects of let-7 on breast cancer through let-7/p53 regulatory loop
p53 was inhibited in let-7 overexpressed cells, we therefore conjectured that the decreased p53 was not induced by let-7 miRNAs, and the increased p53 may help let-7 to repress breast cancer. We added 10 mM of N3 into let-7a/b infected MCF-7 and ZR75-1 cells respectively, and surprisingly, they together showed strong inhibition on cell proliferation (Figure S3b) and self-renewal of stem cells (Figure 2e); with p53 increasing in cancer cells (Figure 3a) and CSCs (Figure 3b).
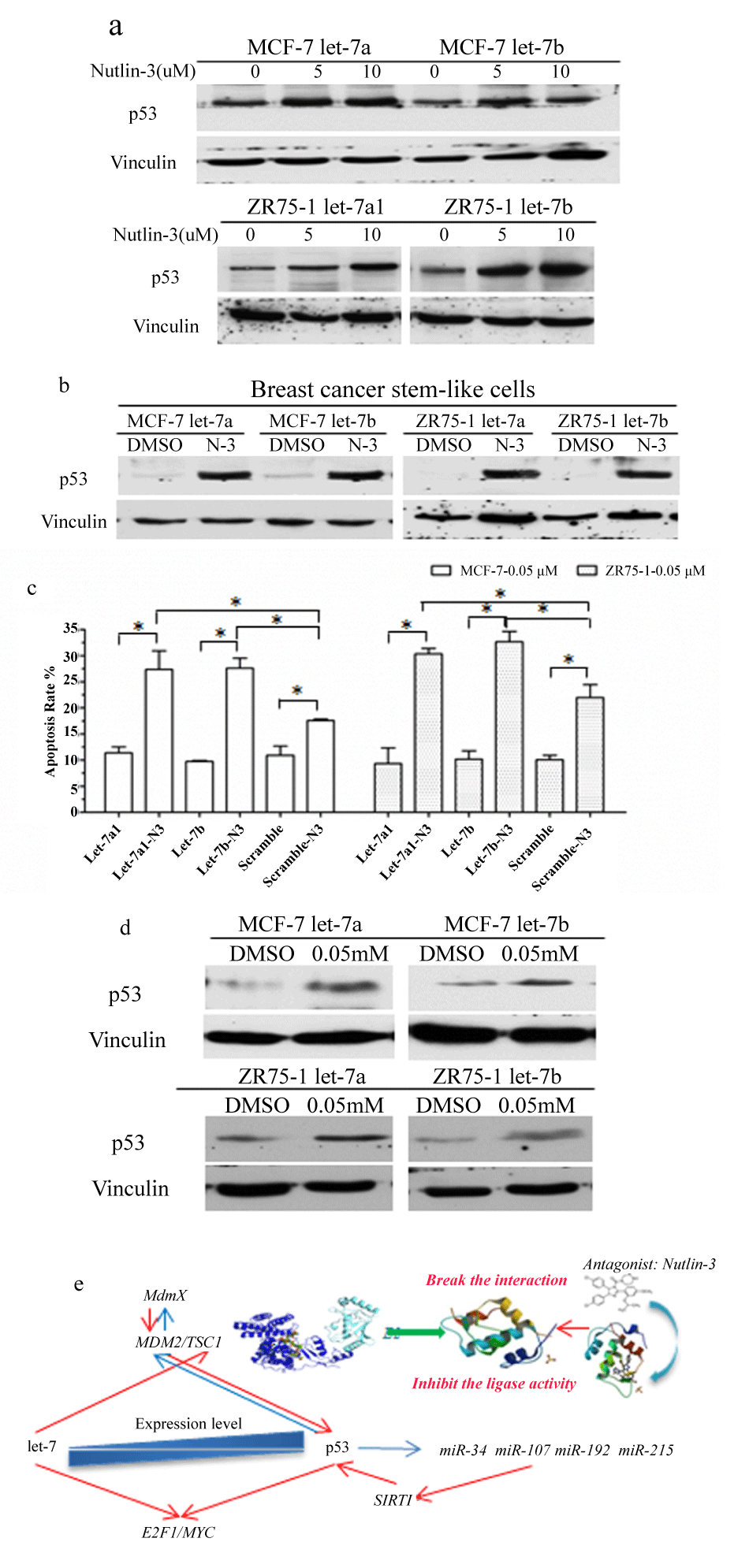
.
Figure 3: The expression levels of let-7 and p53 in response to DNA damage. N3 increased the p53 expression in let-7a/b overexpressed breast cancer cells (a) and mammosphers (b); (c) The let-7a/b over expressed MCF-7 and ZR75-1 cells were treated either with 0.05 mM doxorubicin or combined with N3 for 24 hours and then analyzed for apoptosis, compared with control groups, to identify the functions of over expressed let-7 and p53 on doxorubicin induced cell death. Apoptotic ratios were not significant in let-7a/b increased cells compared to the control group; however, N3 strongly increased let-7 over expressed cell sensitivity to DNA damage inducing agent, p < 0.01; (d) p53 expression level was stimulated by 0.05 mM of doxorubicin; (e) The illustration of let-7/p53 relationships.
View Figure 3
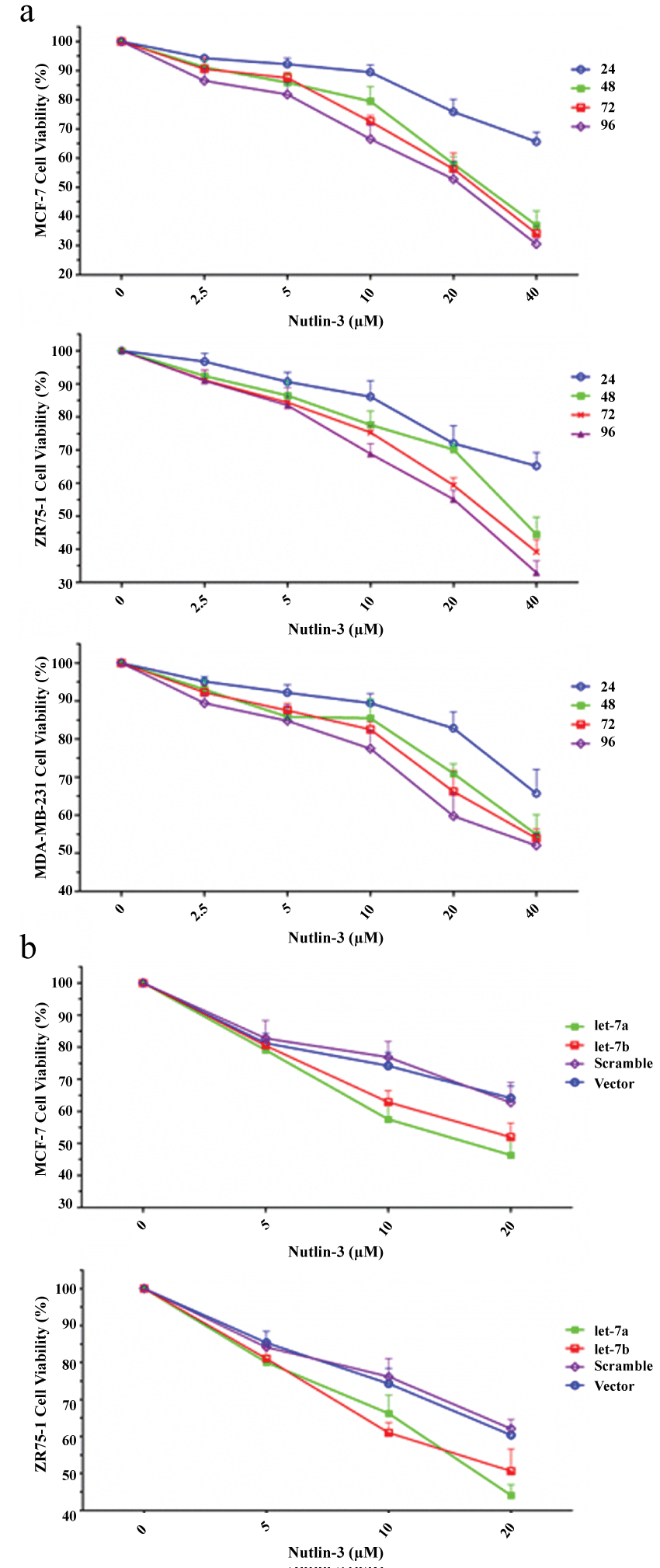
.
Figure S3: The alternative expression levels of let-7 and p53in response toDNA damage. (a) Increased p53 inhibited the proliferation of MCF-7, ZR75-1 and MDA-MB-231 cells significantly (p < 0.01); (b) The increased p53 in let-7a1/b overexpressed MCF-7 and ZR75-1 cancer cells showed strong inhibition on cancer cell proliferation.
View Figure S3
Let-7 was associated with stress signal-induced apoptosis in breast cancer cells
p53 was the key factor in DNA damage response to cell stress signals and could be induced by DNA damage reagents. We then identified if let-7/p53 loop worked in doxorubicin induced cell apoptosis. First, the let-7a/b overexpressed MCF-7 and ZR75-1 cells were treated with 0.05 mM doxorubicin for 24 hours and then analyzed for apoptosis rates, compared with control groups, to determine the functions of let-7 on doxorubicin induced cell death. Apoptotic ratios were not significant in let-7a/b increased cells compared to the control group, p > 0.05 (Figure 3c). To our surprise, N3 strongly increased let-7 overexpressed cell sensitivity to DNA damage inducing agent, with p53 increasing, p < 0.01 (Figure 3d).
Discussions and Conclusions
Let-7 miRNAs are post-transcriptional regulators, controlling cellular biology and cell fates, and lots of oncogenes have been identified to be preys of let-7 and other suppressive miRNAs p [4,18,19]. Cancer recurrence and resistance to chemo-radiotherapy is always the hot potato to our clinicians, and cancer stem cells emerged as the roots of malignancies [20,21]. The therapies aiming at eliminating cancer stem cells are now considered having the most potential and promising new ones in anticancer research [4].
Tumors are groups of heterogeneous cells, containing some silent and resistant stem cells, made for tumor bulk survival through chemo-radiotherapy and to achieve a long-term tumor existence. Strategies specially targeting these CSCs bring hopes to sensitive cancer bulk and to anti-cancer treatments [8,9]. Cancer stem cells show a high ability of self-renewal with specific markers, which could help to isolate the group enriching cancer stem cells, and the cells in mammospheres were demonstrated to act as cancer stem cells [14,22-24]. In this research, we first explored the effects of let-7 miRNAs and p53 on breast cancer cells and BrCSCs, finding that both of them inhibited cancer cell proliferation, and the self-renewal of M1 and M2 spheres. However, it seems they cannot coexist with each other, although they cooperated well. In breast cancer cells increasing let-7 resulted in p53 decrease and vice versa. By treating let-7 overexpressed cancer cells with N3 we identified a stronger inhibition of N3 on breast cancer stem cells. Thus, our research revealed, for the first time that let-7 and p53 inhibited each other in breast cancer cells, forming a negative feedback loop, which may be crucial for sustaining the survival of cancer cells and stem cells (Figure 3e). The suppression of target gene by suppressive miRNAs doesn't always leads to anticancer results. Once the balance was broken with up-regulated wild type p53, the cancer cells and cancer stem cells failed to show malignancy abilities.
Acknowledgements
This experiment was supported by the National Natural Science Foundation of China, approved ID: 81272418. The authors acknowledge assistants in the Institution of Oncology and the staff of the Key Laboratory of Environment and Genes Related to Disease, Ministry of Education, for their technical assistance. All co-authors implicated in this research approved this article to be published.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
-
Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME (2010) The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer 17: F19-36.
-
Liu YM, Xia Y, Dai W, Han HY, Dong YX, et al. (2014) Cholesterol-conjugated let-7a mimics: antitumor efficacy on hepatocellular carcinoma in vitro and in a preclinical orthotopic xenograft model of systemic therapy. BMC Cancer 14: p. 889.
-
Sun X, Jiang S, Liu J, Wang H, Zhang Y, et al. (2015) MiR-208a stimulates the cocktail of SOX2 and β-catenin to inhibit the let-7 induction of self-renewal repression of breast cancer stem cells and formed miR208a/let-7 feedback loop via LIN28 and DICER1. Oncotarget 6: 32944-32954.
-
Yang X, Xie X, Xiao YF, Xie R, Hu CJ, et al. (2015) The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett 360: 119-124.
-
Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, et al. (2003) Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A 100: 10393-10398.
-
Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, et al. (2009) The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 138: 1083-1095.
-
Iglesias-Bartolome R, Patel V, Cotrim A, Leelahavanichkul K, Molinolo AA, et al. (2012) mTOR inhibition prevents epithelial stem cell senescence and protects from radiation-induced mucositis. Cell Stem Cell 11: 401-414.
-
Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597-610.
-
Henle SJ, Wang G, Liang E, Wu M, Poo MM, et al. (2011) Asymmetric PI(3,4,5)P3 and Akt signaling mediates chemotaxis of axonal growth cones. J Neurosci 31: 7016-7027.
-
Hau A, Ceppi P, Peter ME (2012) CD95 is part of a let-7/p53/miR-34 regulatory network. PLoS One 7: e49636.
-
Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T (2008) let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis 29: 2073-2077.
-
Zisoulis DG, Kai ZS, Chang RK, Pasquinelli AE (2012) Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature 486: 541-544.
-
Chatterjee S, Crozet L, Damotte D, Iribarren K, Schramm C, et al. (2014) TLR7 promotes tumor progression, chemotherapy resistance, and poor clinical outcomes in non-small cell lung cancer. Cancer Res 74: 5008-5018.
-
Delort L, Perrier S, Dubois V, Billard H, Mracek T, et al. (2013) Zinc-α2-glycoprotein: a proliferative factor for breast cancer? In vitro study and molecular mechanisms. Oncol Rep 29: 2025-2029.
-
Liu M, Casimiro MC, Wang C, Shirley LA, Jiao X, et al. (2009) p21CIP1 attenuates Ras- and c-Myc-dependent breast tumor epithelial mesenchymal transition and cancer stem cell-like gene expression in vivo. Proc Natl Acad Sci U S A 106: 19035-19039.
-
Taurin S, Sandbo N, Yau DM, Sethakorn N, Dulin NO (2008) Phosphorylation of beta-catenin by PKA promotes ATP-induced proliferation of vascular smooth muscle cells. Am J Physiol Cell Physiol 294: C1169-1174.
-
Chao MW, Chu PC, Chuang HC, et al. (2015) Non-epigenetic function of HDAC8 in regulating breast cancer stem cells by maintaining Notch1 protein stability. Oncotarget .
-
Sun X, Qin S, Fan C, Xu C, Du N, et al. (2013) Let-7: a regulator of the ERα signaling pathway in human breast tumors and breast cancer stem cells. Oncol Rep 29: 2079-2087.
-
Jilkine A, Gutenkunst RN (2014) Effect of dedifferentiation on time to mutation acquisition in stem cell-driven cancers. PLoS Comput Biol 10: e1003481.
-
Sterenczak KA, Eckardt A, Kampmann A, Willenbrock S, Eberle N, et al. (2014) HMGA1 and HMGA2 expression and comparative analyses of HMGA2, Lin28 and let-7 miRNAs in oral squamous cell carcinoma. BMC Cancer 14: 694.
-
Burgess DJ (2014) Stem cells: competitive behaviour of cancer mutations. Nat Rev Cancer 14: 5.
-
Chen G, Umelo IA, Lv S, Teugels E, Fostier K, et al. (2013) miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One 8: e60317.
-
Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, et al. (2008) Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene 27: 5643-5647.
-
Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, et al. (2009) Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res 69: 1279-1283.





