International Journal of Pathology and Clinical Research
Epigenomic Explanations for the Uncertainty of Cancer Biomarkers
Shou-Tung Chen1, Chia-Chen Hsu2, Yu-Wei Leu2 and Shu-Huei Hsiao2*
1Comprehensive Breast Cancer Center, Changhua Christian Hospital, Changhua, Taiwan
2Human Epigenomics Center, Department of Life Science, Institute of Molecular Biology and Institute of Biomedical Science, National Chung Cheng University, Chia-Yi, Taiwan
*Corresponding author:
Shu-Huei Hsiao, PhD, Human Epigenomics Center, Department of Life Science, Institute of Molecular Biology and Institute of Biomedical Science, National Chung Cheng University, 168 University Drive, Min-Hsiung, Chia-Yi, 62102, Taiwan, Tel: 886-5-2720411, 53202, Fax: 886.5.272.2871, E-mail: bioshh@ccu.edu.tw
Int J Pathol Clin Res, IJPCR-2-032, (Volume 2, Issue 2), Review Article; ISSN: 2469-5807
Received: February 10, 2016 | Accepted: March 28, 2016 | Published: April 01, 2016
Citation: Shou-Tung C, Chia-Chen H, Yu-Wei L, Shu-Huei H (2016) Epigenomic Explanations for the Uncertainty of Cancer Biomarkers. Int J Pathol Clin Res 2:032. 10.23937/2469-5807/1510032
Copyright: © 2016 Shou-Tung C, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Inheritable epigenetic modifications, like histone modifications and DNA methylation, were once considered somatically stable and tissue-specific, yet accumulating evidence suggests the contrary. Environmental encounters are transduced into the cell through signaling pathways and these signals are relayed to the nucleus and memorized as epigenetic marks on target genes. Signaling-specific epigenomic changes provide selectable outlines for further lineage determination during differentiation and serve as traceable biomarkers of specific signaling pathways during transformation. Current "big data" interrogation employs signal-specific reduction methods to identify epigenetically modified genes as possible signal targets. These pathway-based analyses show promise for finding true tumor biomarkers, but fail to explain why the onset of cancer can still be partially due to "bad luck". Recent evidence indicates that epigenetically-governed repetitive sequences, especially long and short interspersed nuclear elements (LINE and SINE), in the somatic genome are not evenly distributed. Cell type-specific and even unique single-cell LINE1 transpositions are found in specific brain regions, demonstrating the unevenness of LINE1 in the somatic genome. Retrotransposable elements like LINE1 are silenced, an epigenetic mechanism that stabilizes the genome. Unchained LINE retrotranspositions are found in pre-malignant colon and gastric cancers, and accumulated random transpositions found during the course of cancer development might account for the onset of disease. Epigenetic codes relay and translate cellular encounters into selectable biomarkers and co-evolved with tumorigenesis. Epigenetic regulators are responsible for the maintenance of genomic stability and for the prevention of random transposition. Therefore, changes in epigenetic regulation might explain the probability of oncogenesis and could serve as predictive biomarkers.
Keywords
DNA methylation, Retrotransposon, Signal transduction, Epigenetics
Introduction
With the accumulation of "big data" on genetics in human diseases [1], the likelihood of onset of various cancers is better understood. Identification of causal or associated changes in biomarkers would be impossible if the onset of cancer were entirely stochastic. On the other hand, big data has revealed a degree of uncertainty in the human genome. Even the idea that somatic cells from the same lineage or tissue contain identical genomic material has faded away [2]. Tumorigenesis occurs as a series of minor mutations rather than resulting from a single, high frequency mutation like P53 and APC [3]. Moreover, in some cases mere "misfortune" can be credited as one of the reasons that a patient develops cancer [4,5]. In efforts to explain at least some of the apparent stochasticity in tumorigenesis, there are recent attempts to identify cancer targets or markers associated with cancer. An epigenomic explanation for some of the unpredictability in cancer biomarkers might assist researchers and clinicians in the predictive use of these markers.
Clonal Evolution of Somatic Stem Cells and Cancers
Somatic cells were once believed to possess identical genomic contents since they are all derived from a single cell [6,7]. During development, growth factors and other environmental signals reshape gene expression to generate different lineages of cells that later become specific tissues [8-10]. During this process, external signals are relayed into the cells and are memorized as an inheritable code in the form of specific epigenetic modifications that restrain cell fate [11-13] (Figure 1). The internalization and translation of external signals into epigenetic codes occurs during the early stages of development and germ cell formation but rarely are identified or discussed during somatic stem cell differentiation or tumorigenesis [13-15].
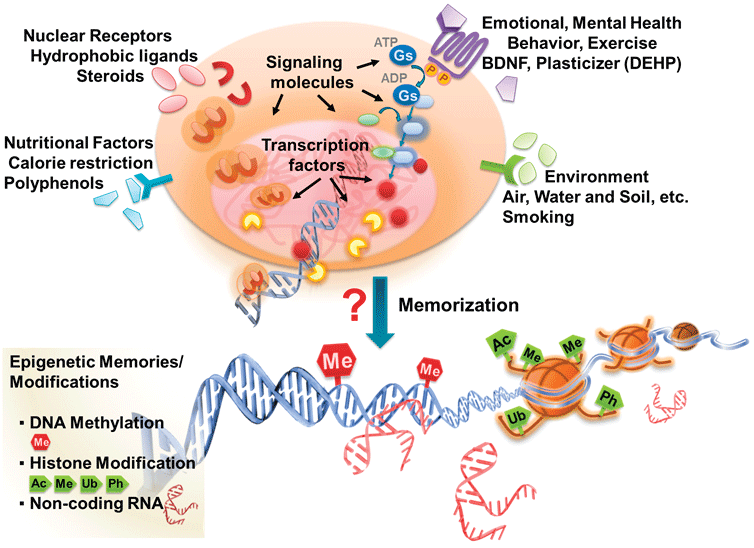
.
Figure 1: Signal-governed epigenomic changes. Environmental cues can be transduced into cells, relayed into nuclei, and memorized as epigenetic marks. The unknown mechanism by which external signals are translated into epigenetic codes is marked by a red question mark.
View Figure 1
The epigenetic code includes DNA methylation, histone modification, and non-coding RNA regulation [16-19]. Specific epigenetic combinations indicate gene expression states as well as cell physiology [20-22]. For example, the Polycomb group proteins EZH2 and YY1 specifically methylate histone H3 at dimethylated lysine 27 (H3K27me2) into the trimethylated form (H3K27me3) [23,24]. This particular histone mark is enriched in stem cells and is associated with stemness [9,13,14,25,26]. The loss of the third methyl group on H3K27 at several genomic loci is sufficient to induce stem cell differentiation [25,27-30]. The same epigenetic mark is found in colon cancer cells but not in differentiated cells [25,31]. This, and similar findings, have been used to support the cancer stem cell theory.
In addition to determination of gene expression patterns, epigenetic modifications are also responsible for genome stability [32,33]. Forty-five to fifty percent of the human genome consists of repetitive sequences [34]. These repetitive sequences may be the evolutionary source of variation. For example, the endogenous retrotransposon Long Interspersed Nuclear Element 1 (LINE1) comprises around 17% of the human genome [35]. Some LINE1 sites are able to transpose autonomously and can induce harmful disruptions in the genome [36-38]. Ancient LINE1s are silenced by a distinct set of epigenetic marks; more recently derived LINE1s are mostly silenced by DNA methylation, suggesting that our genome evolved unevenly [39].
It is clear that somatically inheritable epigenetic modifications must fulfill at least two missions during normal differentiation: one is to generate gene expression differences that restrict cell fate changes; the other is to maintain genomic stability during differentiation. If either of these missions fails, a cell might be transformed. Thus, an epigenomic mechanism must be responsible for the changes in gene expression and stabilization or destabilization of the genome during normal somatic stem cell differentiation or tumorigenesis.
Genome Instability might contribute to Transformation
External signals not only may lead to differentiation or transformation, but also might be responsible for the maintenance of genome stability before and after signal-induced changes in cell physiology [2]. Repetitive sequences like Short or Long Interspersed Nuclear Elements (SINE or LINE, respectively) are the driving forces behind genomic recombination and evolution [34]. Advances in sequencing technology and single-cell genome sequencing have shown that somatic neurons from the same hippocampus region possess different LINE1 insertions (Figure 2). Surprisingly, these variable insertions and transpositions are needed for normal brain function [2,40,41]. The stability of these repetitive elements is maintained by epigenetic modifications like DNA methylation and histone modification [42,43]. External signals thus induce epigenetic modifications that are needed both to change gene expression and to maintain genome stability before and after differentiation.
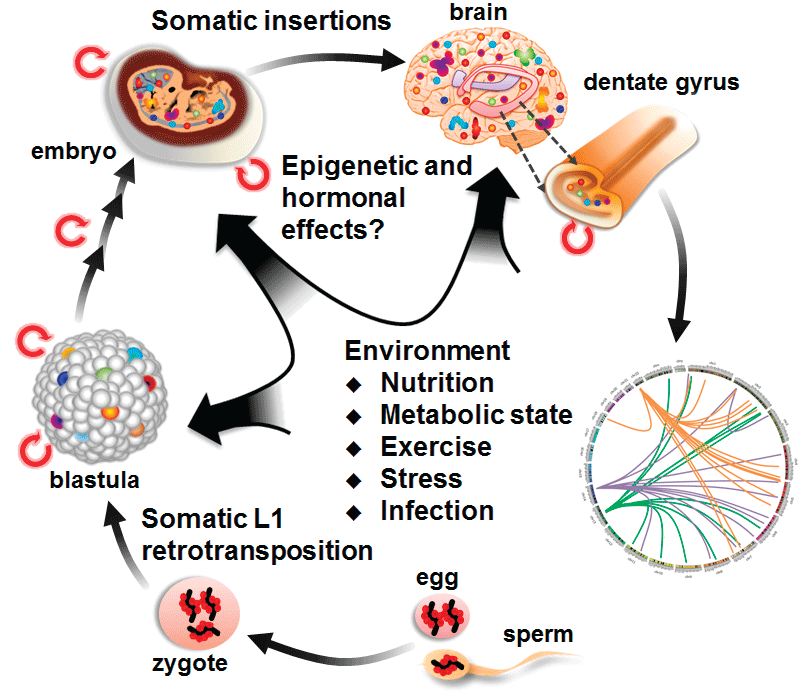
.
Figure 2: During somatic differentiation, the endogenous retrotransposon LINE1 (L1) can transpose if it is not silenced. Individual neurons from different brain regions possess different LINE1 insertions. (Modified from Morgan, G. J., et al., 2012 [52] and Singer, T., et al., 2010 [2]) LINE1 transpositions are represented by the colored lines in Circos plot.
View Figure 2
There are two proposed molecular mechanisms by which abnormal DNA methylation could lead to cancer [44]. One is increased DNA methylation within tumor suppressor genes like HIC1 and RassF1A [15]. The other is loss of global DNA methylation due to low methyltransferase activity that might lead to genome instability and ultimately cancer [45]. The loss of global methylation is hypothesized to mobilize endogenous transposable elements resulting in random insertions, translocations, and transvections [46]. Random transposition could be the "bad luck" element in the onset of cancer [4,41,46]. Indeed, random transposition is documented in gastric and colon cancers [39,47,48]. Targeted sequencing of LINE1 elements shows LINE1 transposition occurring in pre-malignant tissues and progressively accumulating over the four stages of colon and gastric cancers (Figure 3) [39]. The association between LINE1 transposition and the onset and progression of cancer suggests that LINE1 transposition might be one cause of transformation, and furthermore, random transposition might correlate with random onset of cancer [49-51].
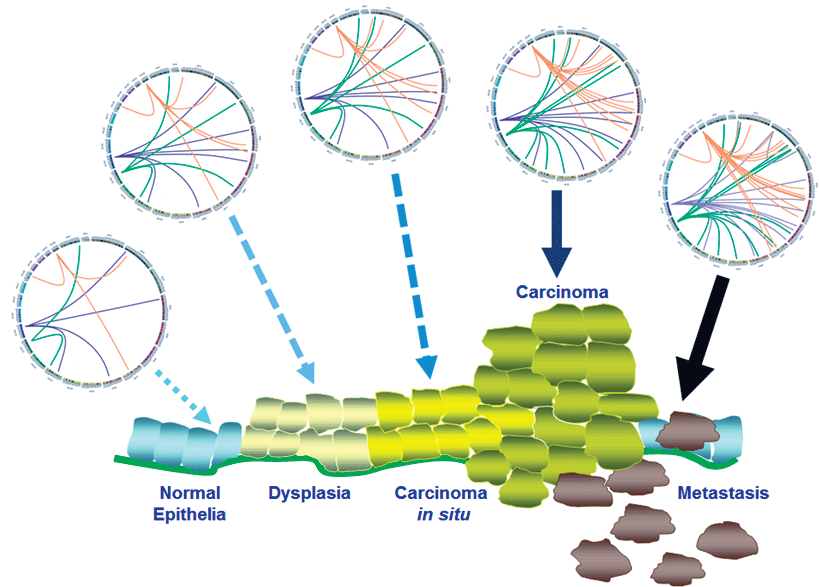
.
Figure 3: Accumulated LINE1 retrotransposition is associated with development of cancers. There are detectable LINE1 retrotransposition in precancer lesions. More and more transposition is observed during tumorigenesis [39]. Increased LINE1 transpositions are represented by the increased colored lines in Circos plot.
View Figure 3
Epigenomic Biomarkers: Signal-Specific Epigenetic Modifications
Environmental signals like exercise, diet, and smoking are transduced into the cell through more than 131 signaling pathways, including Notch, Wnt, BMP, and estrogen receptor (ER) pathways (Figure 1) [52,53]. Changes in a cell's environment can lead to changes in the expression of downstream effector genes and their signal relays [54-57]. Such sequential changes need to be translated into molecular codes via epigenetic modifications to become somatically inheritable [48,58]. Accumulated epigenetic changes then result in varied gene expression and establish selectable parameters for further differentiation (if the changes are "normal") or tumorigenesis (if the changes are "abnormal") [59-61].
The discovery of signal-induced specific epigenetic changes supports the notion that there is internalization of external signals as epigenetic modifications [56,62,63]. For example, if the expression of ER is knocked down, the YY1-governed repression complex initially silences downstream ER targets [56]. Later, DNA methyltransferase (DNMT) adds methyl groups at target loci, and the subsequent passage of cells inherits the methylation pattern [56]. This type of abnormal methylation of the ER target loci Trip10, ENSA, and Casp8AP2 takes place in ER non-expressing breast cancers, proving that the same signal-specific epigenetic modifications occur in vivo [56,64-66].
The assembly of co-regulator complexes has made predicting the downstream epigenome even more complicated. To continue with the same ER example, although ER itself is a nuclear receptor, it teams up with MYC, SMAD, and other transcription factors to up- or down-regulate ER target loci. In cis, MYC and SMAD transcription factor binding sites are located around the ER response element and different bound combinations of these transcription factors produce different patterns of gene expression [60,67,68]. Therefore, cis-regulatory elements and the architecture of the genome are parts of the machinery that programs cell fates [65,69]. Genomic architecture at signaling pathway-regulated sites can make the relay and memorization of signals more efficient. Increasingly, there are blueprints of signal-to-epigenome relays in specific cell types. The results of signals are traceable and the specific patterns of epigenomic changes can reflect the environment around the sequenced cell. Recent research using "big data" has realized the power of reduction studies of the signaling-to-epigenome relay as a blueprint to identify a disease's target genes [70] that may serve as biomarkers.
The Epigenome and Genomic Stability
The three-dimensional (3D) architecture of the genome is critical for genome stability and proper cell physiology [56,71-73]. The genome is organized into regulatory units consisting of multiple loops and inter- and intra-chromosomal interactions [74-76]. With advances in 3C (chromatin conformation capture) and Hi-C techniques [75,77,78], genome-wide maps describing the distribution of these interactions are becoming available. In Caenorhabditis elegans, a recent report indicates that knockout of the genes responsible for sex chromosome dosage compensation can systematically affect genomic interactions within X-chromosome [79]. In contrast, the same mutations do not affect the architecture of autosomes, but still interfere with the expression of autosomal genes [79]. This finding suggests that 3-D conformation, looping, and interactions among genomic regions are specific regulators of gene expression. Nonetheless, how the compartmentalization of regulatory zones is maintained and how compartments can be changed during differentiation or transformation remains unknown.
Understanding how the epigenome, including the methylome, is established, might help to answer these questions. Mammalian cells are separated into germ line and somatic lineages based on their DNA contents. During the early stages of the development, the methylome of these two kinds of cells undergoes drastic revision [80,81]. This methylation process is PIWI-interacting RNA (piRNA)-dependent and may be initiated on repetitive segments within the genome [82]. piRNAs are the largest class of small non-coding RNAs in animal cells, and are clustered within the mammalian genome. Argonaute (Ago) proteins can process their transcripts into pieces of RNAs that are further used to cleave target RNAs into smaller pieces [83,84]. The resulting smaller RNAs in turn process the remaining piRNA cluster transcripts and further fragment the target RNAs [85-88]. This repeated process is named the piRNA "ping-pong" cycle and it is important to amplify the effects of piRNA activity [89].
piRNAs are also crucial for methylation of their target transcripts' promoters [90]. Since piRNAs must enter the nucleus to cause methylation, other factors are needed to direct their translocation and subsequent methylome establishment [91,92]. Although the sequences of piRNAs are not conserved across species, piRNA-mediated silencing targets might be conserved. Indeed, the function of the Arg genes is conserved across species [93,94]. There are eight Arg genes in human cells [95,96]. Recently, researchers observed that Arg proteins need an average of only six nucleotides (nts) to recognize their target RNAs [97]. During the ping-pong amplification cycle, the frame shift between cycle rounds is about 10 nts. Therefore, it is unsurprising that 6-10 nts are needed for Arg proteins to recognize their piRNA interactors. A similar size recognition sequence may be sufficient for piRNA-mediated methylation silencing of targets such as LINE1 retrotransposons [98,99]. In the case of LINE1, 6-10nts is more than enough sequence for target recognition [100,101]. However, the impact of subtle sequence differences between different LINE1 elements, located in different genomic regions, is substantial. Even with only two nucleotide differences, LINE1 regions are sometimes silenced by different epigenetic mechanisms [99,102,103]. Only the most recently evolved LINE1 copies are silenced by DNA methylation; more ancient copies are not [39]. These data suggest that other factors or mechanisms determine the specificity of Arg/piRNA targeting.
Some researchers posit that piRNA clusters are the sequences that capture invading alien DNAs and serve as reservoirs of foreign DNAs information [104,105]. When transcripts with similar sequences are upregulated, piRNA-targeting sequences are able to silence related endogenous retrotransposons or exogenous retrotransposon-like elements [104]. Several constructs with reporter genes have been developed to mimic the capturing system and some of them seemed to be promising until a recent report proved otherwise [106,107]. Using a reporter capturing system, the authors disrupted a piRNA cluster sequence by CRISPR/Cas9 and found that the piRNA targeting system was still functional [108,109]. Since the investigated piRNA targeting system was piRNA cluster-independent, the search for other factors that are involved in the targeting process is necessary to reveal the links between the methylome and genome stability.
Epigenomic Explanations for Uncertainty in Cancer Predictions Using Biomarkers
The relay of the external signals into cells and the memorization of signals as epigenetic marks on target genes are programmed during differentiation and development. No matter how differentiation progresses, genome stability must be maintained to prevent cellular transformation. Thus, the relay of signals and the induction of epigenetic changes are specific, necessary, and under tight control. However, somatic cells are more mosaic than what was previously appreciated. This is evidenced by the extreme example of hippocampal neurons in which LINE1 transpositions occur within individual neurons (Figure 2) [110]. The data suggest that mosaicism is necessary for normal neuronal function [110]. Therefore, there must be a fine line that separates good mosaicism and transposition from deleterious changes. It seems that the pivotal separating point may occur during the relay of the external signals into regulatory epigenetic codes. In normal somatic stem cells the relay system might lead to necessary epigenetic modifications but also to possible randomized mosaicism that decreases genomic stability. Further elucidating the molecular explanations for apparently random epigenomic occurrences will assist researchers in findings biomarkers and other epigenetic identifiers associated with disease onset.
Author Contributions
These authors contributed equally to this work.
Acknowledgement
S-T Chen,Y-W Leu and S-H Hsiao are supported by: NSC-102-2314-B-371-003-MY3. Y-W Leu and S-H Hsiao are supported by:MOST 104-2320-B-194-001.
References
-
Jensen PB, Jensen LJ, Brunak S (2012) Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 13: 395-405.
-
Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH (2010) LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci 33: 345-354.
-
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, et al. (2013) Cancer genome landscapes. Science 339: 1546-1558.
-
Tomasetti C, Vogelstein B (2015) Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347: 78-81.
-
Tomasetti C, Vogelstein B (2015) Cancer risk: role of environment-response. Science 347: 729-731.
-
Brocks D, Assenov Y, Minner S, Bogatyrova O, Simon R, et al. (2014) Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell Rep 8: 798-806.
-
Shackleton M, Quintana E, Fearon ER, Morrison SJ (2009) Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell 138: 822-829.
-
Fernández LP, López-Márquez A, Santisteban P (2015) Thyroid transcription factors in development, differentiation and disease. Nat Rev Endocrinol 11: 29-42.
-
Katoh K, Yamazaki R, Onishi A, Sanuki R, Furukawa T (2012) G9a histone methyltransferase activity in retinal progenitors is essential for proper differentiation and survival of mouse retinal cells. J Neurosci 32: 17658-17670.
-
McLaughlin RN Jr, Young JM, Yang L, Neme R, Wichman HA, et al. (2014) Positive selection and multiple losses of the LINE-1-derived L1TD1 gene in mammals suggest a dual role in genome defense and pluripotency. PLoS Genet 10: e1004531.
-
Hattori N, Niwa T, Kimura K, Helin K, Ushijima T (2013) Visualization of multivalent histone modification in a single cell reveals highly concerted epigenetic changes on differentiation of embryonic stem cells. Nucleic Acids Res 41: 7231-7239.
-
Sreekumar, A., K. Roarty, J.M. Rosen (2015) The mammary stem cell hierarchy: a looking glass into heterogeneous breast cancer landscapes. Endocr Relat Cancer 22: T161-76.
-
Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M (2004) Stem cells and cancer; the polycomb connection. Cell 118: 409-418.
-
Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315-26.
-
Teng IW, Hou PC, Lee KD, Chu PY, Yeh KT, et al. (2011) Targeted methylation of two tumor suppressor genes is sufficient to transform mesenchymal stem cells into cancer stem/initiating cells. Cancer Res 71: 4653-63.
-
Dawson MA, Kouzarides T (2012) Cancer epigenetics: from mechanism to therapy. Cell 150: 12-27.
-
Esteller M (2008) Epigenetics in cancer. N Engl J Med 358: 1148-1159.
-
Feinberg AP, Tycko B (2004) The history of cancer epigenetics. Nat Rev Cancer 4: 143-153.
-
Bird A (2007) Perceptions of epigenetics. Nature 447: 396-398.
-
Smith LT, Otterson GA, Plass C (2007) Unraveling the epigenetic code of cancer for therapy. Trends Genet 23: 449-456.
-
Turner BM (2007) Defining an epigenetic code. Nat Cell Biol 9: 2-6.
-
Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, et al. (2003) Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci 983: 84-100.
-
Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V (2004) The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 18: 2627-2638.
-
Xu CR, Schaffer L, Head SR, Feeney AJ (2008) Reciprocal patterns of methylation of H3K36 and H3K27 on proximal vs. distal IgVH genes are modulated by IL-7 and Pax5. Proc Natl Acad Sci U S A 105: 8685-8690.
-
Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, et al. (2007) Epigenetic stem cell signature in cancer. Nat Genet 39: 157-158.
-
Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, et al. (2009) Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458: 529-533.
-
Kidder BL, Hu G, Zhao K (2014) KDM5B focuses H3K4 methylation near promoters and enhancers during embryonic stem cell self-renewal and differentiation. Genome Biol 15: R32.
-
Jacobs JE, Wagner M, Dhahbi J, Boffelli D, Martin DIK, et al. (2013) Deficiency of MIWI2 (Piwil4) induces mouse erythroleukemia cell differentiation, but has no effect on hematopoiesis in vivo. PLoS One 8: e82573.
-
Weishaupt H, Sigvardsson M, Attema JL (2010) Epigenetic chromatin states uniquely define the developmental plasticity of murine hematopoietic stem cells. Blood 115: 247-256.
-
Xu J, Pope SD, Jazirehi AR, Attema JL, Papathanasiou P, et al. (2007) Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc Natl Acad Sci U S A 104: 12377-12382.
-
Jones PA, Baylin SB (2007) The epigenomics of cancer. Cell 128: 683-692.
-
De La Rosa-Velázquez IA, Rincón-Arano H, Benítez-Bribiesca L, Recillas-Targa F (2007) Epigenetic regulation of the human retinoblastoma tumor suppressor gene promoter by CTCF. Cancer Res 67: 2577-2585.
-
Barreto G, Schäfer A, Marhold J, Stach D, Swaminathan SK, et al. (2007) Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445: 671-675.
-
Cordaux R, Batzer MA (2009) The impact of retrotransposons on human genome evolution. Nat Rev Genet 10: 691-703.
-
Verneau O, Catzeflis F, Furano AV (1998) Determining and dating recent rodent speciation events by using L1 (LINE-1) retrotransposons. Proc Natl Acad Sci USA 95: 11284-11289.
-
Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, et al. (2004) Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene 23: 8841-8846.
-
Furano AV (2000) The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog Nucleic Acid Res Mol Biol 64: 255-294.
-
Hohjoh H, Minakami R, Sakaki Y (1990) Selective cloning and sequence analysis of the human L1 (LINE-1) sequences which transposed in the relatively recent past. Nucleic Acids Res 18: 4099-4104.
-
Ewing AD, Gacita A, Wood LD, Ma F, Xing D, et al. (2015) Wide spread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res 25: 1536-1545.
-
Erwin JA, Marchetto MC, Gage FH (2014) Mobile DNA elements in the generation of diversity and complexity in the brain. Nat Rev Neurosci 15: 497-506.
-
Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, et al. (2009) L1 retrotransposition in human neural progenitor cells. Nature 460: 1127-1131.
-
Suzuki MM, Bird A (2008) DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9: 465-476.
-
Smith ZD, Meissner A (2013) DNA methylation: roles in mammalian development. Nat Rev Genet 14: 204-220.
-
Baylin SB, Ohm JE (2006) Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 6: 107-116.
-
Cadieux B, Ching TT, VandenBerg SR, Costello JF (2006) Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res 66: 8469-8476.
-
Tubio JM, Ju YS, Martincorena I, Cooke SL, Tojo M, et al. (2014) Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 345: 1251343.
-
Hur K, Cejas P, Feliu J, Moreno-Rubio J, Burgos E, et al. (2014) Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 63: 635-646.
-
Ramzy II, Omran DA, Hamad O, Shaker O, Abboud A (2011) Evaluation of serum LINE-1 hypomethylation as a prognostic marker for hepatocellular carcinoma. Arab J Gastroenterol 12: 139-142.
-
Rhee YY, Kim MJ, Bae JM, Koh JM, Cho NY, et al. (2012) Clinical outcomes of patients with microsatellite-unstable colorectal carcinomas depend on L1 methylation level. Ann Surg Oncol 19: 3441-3448.
-
Kato K, Maesawa C, Itabashi T, Fujisawa K, Otsuka K, et al. (2009) DNA hypomethylation at the CpG island is involved in aberrant expression of the L1 cell adhesion molecule gene in colorectal cancer. Int J Oncol 35: 467-476.
-
Menendez L, Benigno BB, McDonald JF (2004) L1 and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Mol Cancer 3: 12.
-
Boyd KD, Pawlyn C, Morgan GJ, Davies FE (2012) Understanding the molecular biology of myeloma and its therapeutic implications. Expert Rev Hematol 5: 603-617.
-
Katoh M (2007) Networking of WNT, FGF, Notch, BMP, and Hedgehog signaling pathways during carcinogenesis. Stem Cell Rev 3: 30-38.
-
Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204-220.
-
Jiang X, Tan J, Li J, Kivimäe S, Yang X, et al. (2008) DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell 13: 529-541.
-
Chuang TC, Moshir S, Garini Y, Chuang AY, Young IT, et al. (2004) The three-dimensional organization of telomeres in the nucleus of mammalian cells. BMC Biol 2: 12.
-
Aithal MG, Rajeswari N (2013) Role of Notch signaling pathway in cancer and its association with DNA methylation. J Genet 92: 667-675.
-
Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL (2006) Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect 114: 567-572.
-
Cho YH, Yazici H, Wu HC, Terry MB, Gonzalez K, et al. (2010) Aberrant promoter hypermethylation and genomic hypomethylation in tumor, adjacent normal tissues and blood from breast cancer patients. Anticancer Res 30: 2489-2496.
-
Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, et al. (2006) Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol Cell 21: 393-404.
-
Feinberg AP, Vogelstein B (1983) Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301: 89-92.
-
Rosenfeld MG, Lunyak VV, Glass CK (2006) Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev 20: 1405-1428.
-
Tang Y, Luo Y, Jiang Z, Ma Y, Lin CJ, et al. (2012) Jak/Stat3 signaling promotes somatic cell reprogramming by epigenetic regulation. Stem Cells 30: 2645-2656.
-
Chen YL, Ko CJ, Lin PY, Chuang WL, Hsu CC, et al. (2012) Clustered DNA methylation changes in polycomb target genes in early-stage liver cancer. Biochem Biophys Res Commun 425: 290-296.
-
Hsiao SH, Huang TH, Leu YW (2009) Excavating relics of DNA methylation changes during the development of neoplasia. Semin Cancer Biol 19: 198-208.
-
Hsiao SH, Lee KD, Hsu CC, Tseng MJ, Jin VX, et al. (2010) DNA methylation of the Trip10 promoter accelerates mesenchymal stem cell lineage determination. Biochem Biophys Res Commun 400: 305-312.
-
Giulianelli S, Vaqué JP, Soldati R, Wargon V, Vanzulli SI, et al. (2012) Estrogen receptor alpha mediates progestin-induced mammary tumor growth by interacting with progesterone receptors at the cyclin D1/MYC promoters. Cancer Res 72: 2416-2427.
-
Paez-Pereda M, Giacomini D, Refojo D, Nagashima AC, Hopfner U, et al. (2003) Involvement of bone morphogenetic protein 4 (BMP-4) in pituitary prolactinoma pathogenesis through a Smad/estrogen receptor crosstalk. Proc Natl Acad Sci U S A 100: 1034-1039.
-
Uhl, JD, Cook TA, Gebelein B (2010) Comparing anterior and posterior Hox complex formation reveals guidelines for predicting cis-regulatory elements. Dev Biol 343: 154-166.
-
Vivar JC, Pemu P, McPherson R, Ghosh S (2013) Redundancy control in pathway databases (ReCiPa): an application for improving gene-set enrichment analysis in Omics studies and "Big data" biology. OMICS 17: 414-422.
-
Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, et al. (2013) A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503: 290-294.
-
Laflamme G, Tremblay-Boudreault T, Roy MA, Andersen P, Bonneil É, et al. (2014) Structural maintenance of chromosome (SMC) proteins link microtubule stability to genome integrity. J Biol Chem 289: 27418-27431.
-
Zilio N, Codlin S, Vashisht AA, Bitton DA, Head SR, et al. (2014) A novel histone deacetylase complex in the control of transcription and genome stability. Mol Cell Biol 34: 3500-3514.
-
Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, et al. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665-1680.
-
Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, et al. (2006) Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38: 1348-1354.
-
Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T (2005) Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet 37: 31-40.
-
Louwers M, Splinter E, van Driel R, de Laat W, Stam M. (2009) Studying physical chromatin interactions in plants using Chromosome Conformation Capture (3C). Nat Protoc 4: 1216-1229.
-
Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, et al. (2015) Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet 47: 598-606.
-
Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, et al. (2015) Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523: 240-244.
-
Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, et al. (2014) DNA methylation dynamics of the human preimplantation embryo. Nature 511: 611-615.
-
Senti KA, Brennecke J (2010) The piRNA pathway: a fly's perspective on the guardian of the genome. Trends Genet 26: 499-509.
-
Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, et al. (2008) DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908-917.
-
Kawaoka S, Hara K, Shoji K, Kobayashi M, Shimada T, et al. (2013) The comprehensive epigenome map of piRNA clusters. Nucleic Acids Res 41: 1581-1590.
-
Oey HM, Youngson NA, Whitelaw E (2011) The characterisation of piRNA-related 19mers in the mouse. BMC Genomics 12: 315.
-
Betel D, Sheridan R, Marks DS, Sander C (2007) Computational analysis of mouse piRNA sequence and biogenesis. PLoS Comput Biol 3: e222.
-
Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ (2012) The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 491: 279-283.
-
Le Thomas A, Marinov GK, Aravin AA (2014) A transgenerational process defines piRNA biogenesis in Drosophila virilis. Cell Rep 8: 1617-1623.
-
Zamore PD (2010) Somatic piRNA biogenesis. EMBO J 29: 3219-3221.
-
Tushir JS, Zamore PD, Zhang Z (2009) SnapShot: mouse piRNAs, PIWI proteins, and the ping-pong cycle. Cell 139: 830-830.
-
Wu D, Fu H, Zhou H, Su J, Zhang F, et al. (2015) Effects of Novel ncRNA Molecules, p15-piRNAs, on the Methylation of DNA and Histone H3 of the CDKN2B Promoter Region in U937 Cells. J Cell Biochem 116: 2744-2754.
-
Sigurdsson MI, Smith AV, Bjornsson HT, Jonsson JJ (2012) The distribution of a germline methylation marker suggests a regional mechanism of LINE-1 silencing by the piRNA-PIWI system. BMC Genet 13: 31.
-
Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, et al. (2008) A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785-799.
-
Zhao J, Sun H, Deng W, Li D, Liu Y, et al. (2010) Piwi-like 2 mediates fibroblast growth factor signaling during gastrulation of zebrafish embryo. Tohoku J Exp Med 222: 63-68.
-
Cox DN, Chao A, Baker J, Chang L, Qiao D, et al. (1998) A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12: 3715-3727.
-
Hutvagner G, Simard MJ (2008) Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9: 22-32.
-
Sasaki T, Shiohama A, Minoshima S, Shimizu N (2003) Identification of eight members of the Argonaute family in the human genome. Genomics 82: 323-330.
-
Nam S, Ryu H, Son WJ, Kim YH, Kim KT, et al. (2014) Mg2+ effect on argonaute and RNA duplex by molecular dynamics and bioinformatics implications. PLoS One 9: e109745.
-
Manakov SA, Pezic D, Marinov GK, Pastor WA, Sachidanandam R, et al. (2015) MIWI2 and MILI Have Differential Effects on piRNA Biogenesis and DNA Methylation. Cell Rep 12: 1234-1243.
-
Pezic D, Manakov SA, Sachidanandam R, Aravin AA (2014) piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev 28: 1410-1428.
-
Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, et al. (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089-1103.
-
Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, et al. (2007) A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science 315: 1587-1590.
-
Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, et al. (2011) Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480: 264-267.
-
De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, et al. (2011) The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480: 259-263.
-
Yamamoto Y, Watanabe T, Hoki Y, Shirane K, Li Y, et al. (2013) Targeted gene silencing in mouse germ cells by insertion of a homologous DNA into a piRNA generating locus. Genome Res 23: 292-299.
-
Rosenkranz D (2016) piRNA cluster database: a web resource for piRNA producing loci. Nucleic Acids Res 44: D223-230.
-
Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, et al. (2013) Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27: 390-399.
-
Yin H, Lin H (2007) An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450: 304-308.
-
Kumar MS, Chen KC (2012) Evolution of animal Piwi-interacting RNAs and prokaryotic CRISPRs. Brief Funct Genomics 11: 277-288.
-
Ishizu H, Iwasaki YW, Hirakata S, Ozaki H, Iwasaki W, et al. (2015) Somatic Primary piRNA Biogenesis Driven by cis-Acting RNA Elements and trans-Acting Yb. Cell Rep 12: 429-440.
-
Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, et al. (2012) Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 151: 483-496.





