International Journal of Respiratory and Pulmonary Medicine
A Novel Approach to Investigate Functional Exonic SNPs Associated with Lung Diseases at Post-Transcriptional Stage
Tong Zhou and Ting Wang*
Arizona Respiratory Center and Department of Medicine, University of Arizona, USA
*Corresponding author: Dr. Tong Zhou, Research Assistant Professor in Clinical Bioinformatics
Arizona Respiratory Center and Department of Medicine, University of Arizona, Tucson, USA, Tel: 1-520-626-8433, E-mail: tongzhou@email.arizona.edu
Int J Respir Pulm Med, IJRPM-1-002, (Volume 1, Issue 1), Editorial; ISSN: 2378-3516
Received: August 14, 2014 | Accepted: August 16, 2014 | Published: August 18, 2014
Citation: Zhou T, Wang T (2014) A Novel Approach to Investigate Functional Exonic SNPs Associated with Lung Diseases at Post-Transcriptional Stage. Int J Respir Pulm Med 1:002. 10.23937/2378-3516/1410002
Copyright: © 2014 Zhou T. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Genome wide association studies (GWAS) have resulted in the identification of novel genetic loci associated with a variety of diseases and clinical phenotypes, including complex lung disease such as asthma and COPD. However, the translational utilization of these expensive datasets has been restricted by multiple reasons including limited methodology to understand the exact molecular function of these single nucleotide polymorphisms (SNPs).
mRNA encodes amino acid information with linear nucleotide sequences. In addition to mRNA primary sequence, base pairing of nucleotides in mRNAs creates specific secondary structures, such as hairpins and stem-loops. A growing body of studies indicate that mRNA secondary structure contains key regulatory information in different biological processes [1-3], including pre-mRNA splicing [4], microRNA (miRNA) mediated gene regulation [5-7], and protein synthesis [8-11]. Experimental profiling of mRNA structure at genome-scale both in vitro [10,12-14] and in vivo [15] has confirmed the post-transcriptional regulatory role of mRNA secondary structure in various organisms.
SNPs are single base-pair substitutions which occur within genome. SNPs within mRNA can be categorized into three groups by location and function: non-coding SNPs located in 5 untranslated region (5 UTR) and 3' untranslated region (3 UTR), non-synonymous coding SNPs which change protein primary sequences, and synonymous coding SNPs which dont modify the encoded amino acid. The non-synonymous SNPs within mRNAs have been heavily documented and investigated in the past thirty years because this kind of nucleotide substitution potentially alters protein structure and function due to the different chemical/physical properties of the altered amino acid. Nevertheless, at mRNA level, it should be noted that SNPs may also give rise to different allelic forms of mRNA secondary structure, no matter the SNPs are non-synonymous or not, which may consequently affect final protein expression [16-19]. Several SNPs within mRNA are already known to contribute to human diseases by altering mRNA secondary structure [16,19,20]. Comparison of mRNA structure against published GWAS results indicates that some strong genetic signals from GWAS may exhibit their functional effects through modified mRNA structures [18]. However, as of now, the structural effects on mRNA by SNPs have not been systematically analyzed. Especially, the influence of SNPs on protein expression via mRNA structural alteration is largely unknown. Thereby we hypothesize here that a considerable proportion of lung diseaseassociated exonic SNPs may change mRNA secondary structure and in turn affect protein expression (Figure 1).
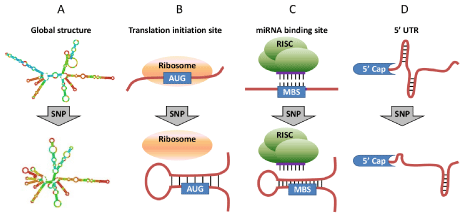 Figure 1: Structural effect caused by exonic SNPs. (A) Exonic SNPs that cause substantial change in mRNA global structure and stability. (B) Exonic SNPs that change the mRNA local structure around translation initiation sites. (C) Exonic SNPs that change the structural accessibility of miRNA binding sites (MBSs). (D)
Exonic SNPs in 5 UTR that may change mRNA local structure near 5 cap and thus affect miRNA-mediated translation inhibition.
View Figure 1
Figure 1: Structural effect caused by exonic SNPs. (A) Exonic SNPs that cause substantial change in mRNA global structure and stability. (B) Exonic SNPs that change the mRNA local structure around translation initiation sites. (C) Exonic SNPs that change the structural accessibility of miRNA binding sites (MBSs). (D)
Exonic SNPs in 5 UTR that may change mRNA local structure near 5 cap and thus affect miRNA-mediated translation inhibition.
View Figure 1
Recently, we investigated a coding no synonymous SNP in the gene of myosin light chain kinase (MYLK), which is associated with severe asthma in African Americans. We have found that this single SNP mediates significant MYLK mRNA structure disruption (not only local effects), thereby inducing mRNA stability alteration and decay rate change. More importantly, the change of global structure of MYLK mRNA by this SNP alters the local accessibility of mRNA translation initiation site, which mediates the change of translational efficiency of the gene. As a first time, it proved that this asthma susceptible non-synonymous SNP changes protein expression levels, while not in the traditional concept of protein activity/structure modification. These novel theories has been further validated , in vitro and in vivo, that, this SNP alters the expression levels of MYLK coded proteins, due to mRNA stability and translation initiation efficiency, and thereby mediate higher susceptibility of severe asthma. This study provides a example of this novel translational approach.
In conclusion, novel methods are needed for these fast-growing GWAS and expression array datasets for lung diseases, especially for the complex diseases in need of more effective and selective therapies. The investigation of the impact of mRNA structural by disease associated SNPs will reveal novel mechanisms of genetic function of SNPs on gene expression and disease susceptibility, therefore identify novel therapeutic targets.
References
-
Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY (2011) Understanding the transcriptome through RNA structure. Nat Rev Genet 12: 641-655.
-
Shen LX, Basilion J P, Stanton VP (1999) Single-nucleotide polymorphisms can cause different structural folds of mRNA. Proc Natl Acad Sci U S A 96: 7871-7876.
-
Gu W,Li M, Xu Y, Wang T, Ko JH, et al (2014) The impact of RNA structure on coding sequence evolution in both bacteria and eukaryotes. BMC Evol Biol 14: 87.
-
Warf M B, Berglund JA (2010) Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci 35: 169-178.
-
Gu W, Wang X, Zhai C, Xie X, Zhou T (2012) Selection on Synonymous Sites for Increased Accessibility around miRNA Binding Sites in Plants. Mol Biol Evol 29: 3037-3044.
-
Meijer HA, Kong YW, Lu WT, Wilcznska A, Spriggs RV, et al.(2013) Translational Repression and eIF4A2 Activity Are Critical for MicroRNA-Mediated Gene Regulation. Science 340: 82-85.
-
Gu W, Xu Y, Xie X, Wang T, Ko JH,et al. (2014) The role of RNA structure at 5' untranslated region in microRNA-mediated gene regulation. RNA.
-
Kozak M (2005) Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361: 13-37.
-
Gu W, Zhou T, Wilke CO (2010) A Universal Trend of Reduced mRNA Stability near the Translation-Initiation Site in Prokaryotes and Eukaryotes. PLoS Comput Bio l6: e1000664.
-
Li F, Zheng Q, Vandivier LE, Willmann MR, Chen Y, et al. (2012) Regulatory Impact of RNA Secondary Structure across the Arabidopsis Transcriptome. Plant Cell 24: 4346-4359.
-
Zhou T, Wilke CO (2011) Reduced stability of mRNA secondary structure near the translation-initiation site in dsDNA viruses. BMC Evol Biol 11: 59.
-
Kertesz M (2010) Genome-wide measurement of RNA secondary structure in yeast. Nature 467: 103-107.
-
Zheng Q, Ryvkin P, Li F, Dragomiri I, Valladares O, et al. (2010) Genome-Wide Double-Stranded RNA Sequencing Reveals the Functional Significance of Base-Paired RNAs in Arabidopsis. PLoS Genet 6: e1001141.
-
Wan Y, Qu K, Ouyang Z, Kertesz M, Li J, Tibshirani R, et al (2012) Genome-wide Measurement of RNA Folding Energies. Mol cell 48: 169-181.
- 15. Ding, Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, et al. (2013) In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Naturead 30: 505.
-
Zhang Y, Wang D, Johnson AD, Papp A C, Sadee W (2005) Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem 280: 32618-32624.
-
Shabalina SA, Zaykin DV, Gris P, Oqurtsov AY, Gauthier J, et al. (2009) Expansion of the human mu-opioid receptor gene architecture: novel functional variants. Hum Mol Genet 18: 1037-1051.
-
Johnson AD, Trumbower H, Sadee W (2011) RNA structures affected by single nucleotide polymorphisms in transcribed regions of the human genome. WebmedCentral BIOINFORMATICS 2.
-
Johnson AD, Wang D, Sadee W (2005) Polymorphisms affecting gene regulation and mRNA processing: broad implications for pharmacogenetics. Pharmacol Ther 106: 19-38.
-
Wang D, Johnson AD, Papp AC, Kroetz DL, Sadee W (2005) Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet Genomics15: 693-704.





