International Journal of Respiratory and Pulmonary Medicine
Distinct Sex-Specific Gene Expression Changes in the Placenta in Association with Childhood Allergy
Astrud R Tuck1,Luke E Grzeskowiak1, Annette Osei-Kumah1, Zarqa Saif2, Suzanne M Edwards3, Andrew Tai4, Susan L Prescott5,8, MeriTulic6,8, Richard Saffery7,8 and Vicki L Clifton1,2,8*
1Robinson Institute, School of Paediatrics and Reproductive Health, University of Adelaide, Australia
2Mater Medical Research Institute, University of Queensland, Australia
3Data Management and Analysis Centre, University of Adelaide, Australia
4Respiratory and Sleep Department, Women's and Children's Hospital, Australia
5School of Paediatrics and Child Health, University of Western Australia, Australia
6University of Nice Sophia-Antipolis, France
7Cancer and Disease Epigenetics, Murdoch Children's Research Institute, Department of Paediatrics, University of Melbourne, Australia
8International Inflammation (in-FLAME) Network, of the World Universities Network (WUN), Australia
*Corresponding author: Professor Vicki L Clifton, Mater Medical Research Institute, Level 4, Translational Research Institute, 37 Kent St, Woolloongabba, Qld. 4101, Brisbane, Australia, Tel: 617-3447640, E-mail: Vicki.clifton@mater.uq.edu.au
Int J Respir Pulm Med, IJRPM-2-033, (Volume 2, Issue 4), Research Article; ISSN: 2378-3516
Received: April 16, 2015 | Accepted: October 27, 2015 | Published: October 29, 2015
Citation: Tuck AR, Grzeskowiak LE, Osei-Kumah A, Saif Z, Edwards SM, et al. (2015) Distinct Sex-Specific Gene Expression Changes in the Placenta in Association with Childhood Allergy. Int J Respir Pulm Med 2:033. 10.23937/2378-3516/1410033
Copyright: © 2015 Tuck AR, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: The prevalence of allergic disease has risen significantly during recent years. A major component of the susceptibility to allergic disease is determined in prenatal life, when the placenta plays a central role in fetal growth and development. In this study, we aimed to identify the patterns of gene expression in the placenta that may program early immune function to increase susceptibility to allergy.
Methods: A set of immune genes known to be associated with asthma, allergy and inflammation were selected for analysis by quantitative real-time polymerase chain reaction (qRT-PCR) on placental tissue from infants who did or did not develop an allergy by 2 years of age. Analysis was performed on males and females separately for each allergy type including eczema, rhinitis or asthma.
Results: Of 11 candidate allergy-associated genes tested by qRT-PCR, 4 were found to be associated with the development of specific childhood allergy types (P < 0.05). These included MMP9 for both males and females that developed eczema, TLR7 for females that developed eczema, KITL1 for males that developed rhinitis and ORMDL3 for females that developed asthma.
Conclusions: This study has identified altered expression of placental genes involved in inflammation in association with the development of specific allergies in childhood. The current data provide supporting evidence implicating the placenta in programming the fetal immune system in early life.
Keywords
Human, Placenta, Allergy, Child, Gene, Immune, Eczema, Rhinitis, Asthma
Introduction
There has been an epidemic rise in allergic disease since the second-half of the twentieth century, particularly in Western countries [1,2]. Diseases such as eczema, allergic rhinitis and food allergies now represent significant burdens to human health. Australia has one of the highest rates of allergic disease, with allergic sensitisation evident in up to 40% of children and asthma accountable as the most common cause of chronic disease in childhood [3,4]. It is now clear that developmental events play a critical role in determining susceptibility to allergy [5-7]. Exposures during pregnancy, when developing fetal systems are particularly vulnerable to environmental influences, could have significant effects on the programming of disease susceptibility [8].
The placenta plays a central role in fetal growth and development and acts as the immunological and metabolic interface between the mother and fetus. It has been demonstrated that levels of immune mediators detected in the fetus, of which the placenta is the major source, correlate with subsequent development of allergy [9]. Furthermore, reduced mRNA levels of regulatory T cell (TREG) marker, FOXP3, have been reported in the placentae of infants who subsequently developed an allergic disease [10]. Thus, the placenta has great potential to program the fetal immune system, potentially increasing the susceptibility to allergy after birth. The concept of altered placental gene expression influencing fetal programming is supported by a recent study in mice. Knockout of the placenta-specific insulin-like growth factor-2 (Igf2) P0 transcript (Igf2-P0 KO) created an imbalance between fetal nutritional demand and placental supply of nutrients, leading to the offspring displaying significantly increased anxiety in later life [11]. Defining the molecular mechanisms in the placenta that may be altered in association with childhood allergy is vital for identifying causal pathways of altered fetal immune programming that increase the risk of allergy susceptibility.
Previous work by our team has demonstrated that the human placenta functions in a sex specific manner with significant differences in global gene and protein expression [12]. In particular there are significant differences in the placental immune response to the presence of maternal allergy [13] and its regulation by glucocorticoids between male and female placentae [14] with female placentae appearing more sensitive to an immune challenge and glucocorticoids relative to male placentae. These findings suggest that any placental immune dysregulation that leads to the susceptibility to allergy in later life may be vary in a sex specific manner.
In this study, we hypothesised that childhood susceptibility to allergy is increased by significant alterations in the in utero environment which include sex specific alterations in placental function that may program the development of the fetal immune system. We aimed to identify genetic pathways altered in the placentae of children who subsequently developed allergy in early childhood and to explore whether there are differences in relation to the allergic phenotype a child develops. We also aimed to determine whether there are sex differences in the expression profile. To do this, we selected a set of genes with immune functions known to be associated with asthma, allergy and inflammation for analysis on placental tissue from male or female infants who did or did not develop an allergy in early childhood. While the immune genes examined in this study were not exhaustive of those previously associated with allergy, we have chosen a select number of strong candidates for investigation in this study based on preliminary findings of placental microarray work comparing children with and without allergy by 4 years of age. From these, we have identified several immune genes that are associated with the development of specific childhood allergy phenotypes.
Methods and Materials
Study participants
This work was approved by the Government of South Australia (SA) Health Human Research Ethics Committee (TQEH/LMH/MH) and the University of Adelaide Human Research Ethics Committee (2009045). Women were recruited from the antenatal clinic at the Lyell McEwin Hospital (Elizabeth, South Australia, Australia) during the first trimester of their pregnancy following written, informed consent. Women who smoked during pregnancy were excluded. Women were assessed throughout pregnancy at 12, 18, 30 and 36 weeks for atopic status, diet and fetal growth. Placentae were collected within 45 minutes of delivery and multiple samples from the central and peripheral regions of the maternal side of placenta were pooled together, snap frozen and stored at -80°C.
Follow-up data was collected on infants every 12 months from 6 months of age until 36 months of age. Allergy (asthma, rhinitis and eczema, but not food allergy) was determined by their general practitioner and/or allergy specialist and reported to the parent, who then completed a modified version of the International Study of Asthma and Allergy in Childhood (ISAAC) questionnaire [15,16], delivered at each follow-up visit. ISAAC is a collaborative project which has developed a standardized methodology to describe the prevalence and severity of asthma, rhinitis and eczema in children throughout the world [15,16]. Key questions from the ISAAC questionnaire were used to gather data on symptoms of asthma, allergic rhinitis, and atopic eczema. Rhinitis was defined if the parents reported "Yes" to the question, "Has your child ever had hayfever?", or if parents responded yes to both questions, "In the last 12 months, has your child had a problem with sneezing, or a runny, or a blocked nose when he/she DID NOT have a cold or the flu?" and "In the last 12 months, has this nose problem been accompanied by itchy/watery eyes?". Eczema was defined if the parents reported "Yes" to any one of the following questions: "In the last 12 months, has your child had a dry itchy rash at any time?" and "Has your child ever had eczema?" Asthma was defined at the 36 month visit if the parents reported "Yes" to the question, "Has your doctor ever told you that your child has asthma?".
RNA extraction
Total RNA was extracted from frozen placental tissue using the Trizol method as previously described [14,17]. RNA concentration was measured using an IMPLEN spectrophotometer, and RNA quality and integrity was assessed using a 2100 Bio analyser (Agilent Technologies, Santa Clara, CA, USA). Only RNA samples demonstrating intact 28S and 28S rRNA peaks and a high RNA integrity number (RIN) (> 6) were included in the study. Reverse transcription was performed on 1 μg of total RNA using the iScript cDNA Synthesis kit (Biorad, Hercules, CA, USA) according to the manufacturer's instructions.
Quantitative RT-PCR
Taqman Gene Expression Assays (Life Technologies, Carlsbad, CA, USA) were used to measure expression of putative allergy-associated genes (identified following microarray analysis, data not included as it was a preliminary assessment) in male (n = 28) and female (n = 26) placentae. Target genes are listed in table 2 and figures 1 and figure 2. qRT-PCR was performed using the 7500 Fast Real-Time PCR System (Applied Biosystems, Mulgrave, VIC, Australia) according the manufacturer's instructions, and two technical replicates were included for each sample per assay. PCR data was analysed using the 2-ΔΔCt method, with expression of each gene normalised to β - actin expression as used previously for gene expression analysis of placental tissue [18,19].
Statistical analysis
Statistical analyses were performed with SPSS Statistics software (version 20, Statistical Package for Social Sciences, SPSS Inc, IBM, Chicago, IL). All data are expressed as median unless otherwise indicated. Sex differences in gene expression between individuals with a specific allergy type relative to individuals without an allergy were tested using non-parametric Mann-Whitney U tests. Non-parametric Kruskal Wallace analysis of variance (KW-ANOVA) was used to compare all groups by sex and allergy phenotype. The impact of maternal asthma on placental gene expression was also examined using Mann-Whitney U tests to determine whether any changes observed in the placenta may be related to the presence of maternal disease.
Results
Characteristics of study populations
Maternal characteristics between "no allergy" and "allergy" study groups were not significantly different. The mean age and BMI of mothers in the "no allergy" group was 29.8 years of age and 25.7, while the mean age was 27.1 years and mean BMI was 28.3 in the "allergy" group. Neonatal characteristics are summarised in supplementary table 1. Maternal and neonatal characteristics of each study group are summarised in table 1.
![]()
Table 1: Maternal characteristics during pregnancy and neonatal characteristics
View Table 1
Quantitative RT-PCR of selected genes
Some placental genes identified from the preliminary microarray work that were selected for analysis were not found to vary significantly in relation to allergic phenotype and so have been listed in table 2.
![]()
Table 2: Inflammatory genes selected for analysis in the placenta that were not altered in relation to childhood allergy.
View Table 2
Placental genes altered in association with childhood eczema
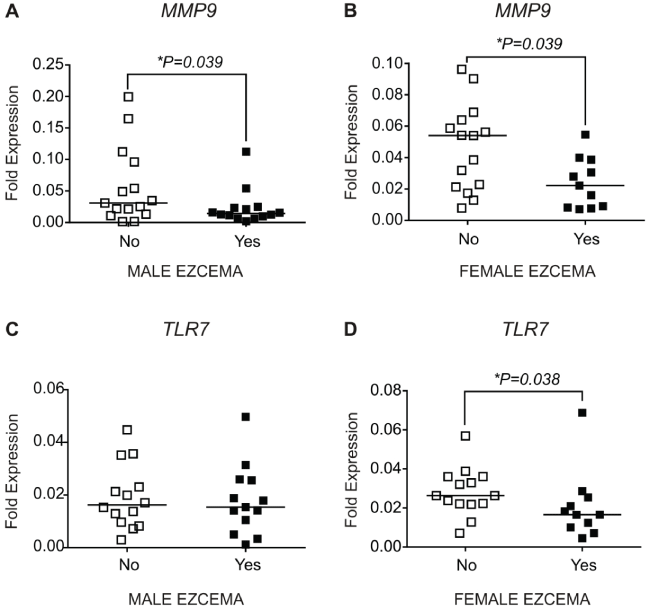
Expression of matrix metallopeptidase 9 (MMP9) mRNA was reduced in placentae from males and females with eczema compared to placentae from males females without eczema (Mann Whitney U test, males: P = 0.039, females: P = 0.039) (Figure 1A and Figure 1B). Overall, MMP9 levels were significantly higher in males than females in subjects with eczema (KW-ANOVA, P = 0.048, data not shown) Levels of Toll-like receptor 7 (TLR7) did not differ in males with or without eczema (Figure 1C), but was lower in placentae of females with eczema compared to placentae of females without eczema (Figure 1D).
.
Figure 1: Gene expression levels in placentae of offspring with eczema compared to placentae from offspring without eczema. The solid bar indicates the median. Genes are expressed relative to β - actin. Male no eczema n = 14, male eczema n = 14, female no eczema n = 15, female eczema n = 11. *indicates significantly different to no allergy, P < 0.05 (Mann-Whitney U test).
View Figure 1
Placental genes altered in association with childhood rhinitis
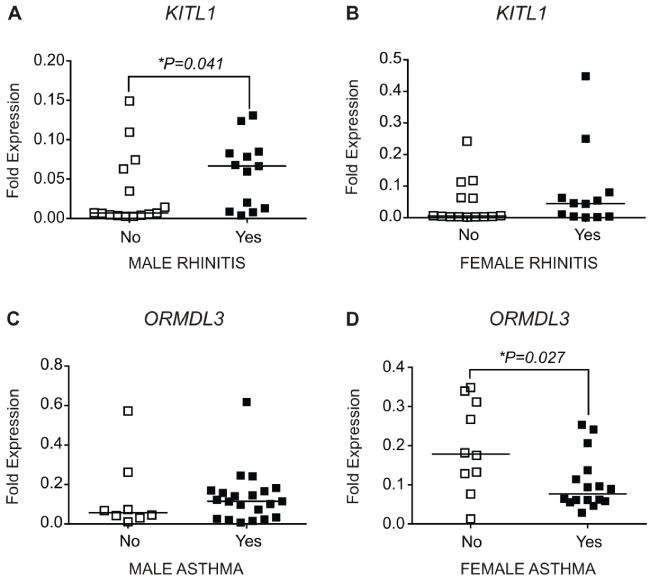
Levels of kit ligand 2 (KITL2) mRNA were very low or absent with Cq values over 35, and thus was not included in statistical analyses. Kit ligand 1 (KITL1) expression was significantly higher in placentae from males with rhinitis compared to placentae from males without rhinitis (P = 0.041) (Figure 2A), while expression did not differ between placentae from females with or without rhinitis (Figure 2B).
.
Figure 2: Gene expression levels in placentae of offspring with either rhinitis or asthma compared to placentae from offspring without rhinitis or asthma. The solid bar indicates the median. Genes are expressed relative to β - actin. Male no rhinitis n = 15, male rhinitis n = 13, female no rhinitis n = 14, female rhinitis n = 12, male no asthma n = 7, male asthma n = 21, female no asthma n = 10, female asthma n = 16. *indicates significantly different to no allergy, P < 0.05 (Mann-Whitney U test).
View Figure 2
Placental genes altered in association with childhood asthma
Expression of ORM (yeast)-like protein isoforms 3 (ORMDL3) mRNA did not differ between the placentae of males with or without asthma, but was reduced in the placentae of females with asthma compared to the placentae of females without asthma (P = 0.027) (Figure 2D).
Relationship of placental gene expression to maternal asthma during pregnancy
We assessed whether there was any relationship between maternal asthma and changes in gene expression, as asthma is a significant risk factor that increases the likelihood of offspring developing allergy [20]. As shown in supplementary table 2, the presence or absence of maternal asthma was not significantly associated with any of the placental genes identified to be associated with childhood allergy.
Discussion
In this study, we have shown that a number of genes expressed in the placenta at birth are differentially expressed in association with the subsequent development of childhood eczema, rhinitis and asthma. This data supports the current hypothesis that in utero events may pre-program the fetal immune system to an allergic phenotype [21]. Our results indicate that placentae of children who subsequently develop allergy have a bias towards a Th2 immune profile which varied sex specifically. These data suggest that both sex-specific and gene-specific mechanisms may underlie the development of each allergy.
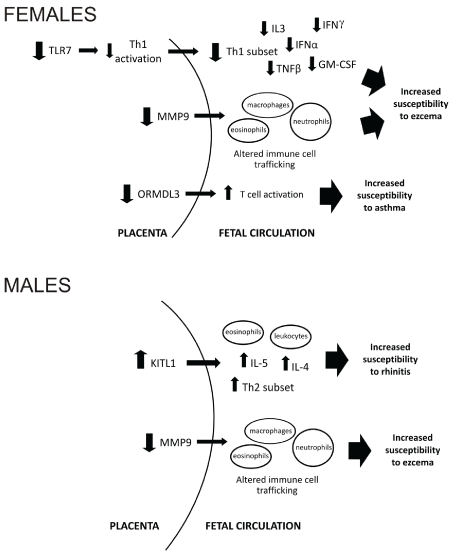
We identified two genes associated with the development of eczema, specifically MMP9 in males and TLR7 in females. TLR7 is an X chromosome gene which does not escape X inactivation; however, it has been suggested that epigenetic mechanisms may cause altered expression between males and females. TLR7 promotes a Th1-type immune response via differentiation of Th1 cells and production of cytokines, and strong evidence indicates that it is protective against the development and severity of allergic disorders [22-25]. Decreased mRNA in placentae from females who subsequently developed eczema may drive decreased placental Th1 cytokine activation including IFNγ, IFNα, TNFβ, IL-3 and GM-CSF [22-25] (summary in Figure 3). A reduction in these pro-inflammatory cytokines may influence T cell differentiation including CD8+ cells [26], T regulatory cells [27] and dendritic cells in the fetus [28], thereby increasing susceptibility to developing eczema.
.
Figure 3: Proposed model of mechanisms underlying increased susceptibility to specific allergy types in females and males.
View Figure 3
There may also be a bias towards a Th2 response in male eczema via different mechanisms. MMP9 plays a significant role in inflammation by facilitating cellular traffic, including neutrophils and eosinophils, via degradation of extracellular matrix and establishment of chemokine gradients [29]. Importantly, upregulation of MMP9 mRNA has been demonstrated to be concomitant with the recruitment of Th2 cells in mice [29], while MMP9-null mice showed a marked attenuation of the Th2 inflammatory response [30]. Altered expression of MMP9 in placentae may result in altered immune cell trafficking and increased Th2 activation, including elevated numbers of CD4+ cells and release of inflammatory cytokines leading to increased susceptibility to developing eczema in male offspring (Figure 3).
This study has shown an association between increased KITL1 expression in male placentae and subsequent development of rhinitis, supporting mouse studies demonstrating KITL to be involved in the development of allergic inflammation [31-33]. We found no differences in expression of the KITL receptor, c-kit, suggesting that any influences of the KITL/c-kit signalling pathway on the programming of the fetal immune system are exerted through elevated KITL expression alone. Abnormal KITL1 gene expression may contribute towards a skewed Th2 immune profile by promoting infiltration of inflammatory cells including eosinophils and leukocytes, and increased production of Th2 cytokines IL-4 and IL-5 [33] as shown in figure 4.
Interestingly, this study identified decreased expression of ORMDL3 to be associated with the development of asthma in females. To our knowledge, no study has characterised the association of ORMDL3 with asthma by sex; thus, this is the first study to demonstrate a sex-specific association between ORMDL3 and asthma. Increased ORMDL3 has recently been identified as being strongly associated with both childhood and adult asthma [34-42]. Therefore, it is somewhat surprising that our study has observed decreased expression in association with asthma. The mechanisms underlying ORMDL3 function and its effects on asthma are unclear and studies have reported conflicting results. Notably, an in vitro study demonstrated that ORMDL3 reduced T-cell activation via an important immune activation mechanism [43]. Overexpression of ORMDL3 in a T-cell line inhibited store-operated Ca2+ influx, thereby reducing nuclear translocation of the nuclear factor of activated T-cells (NFAT) [43]. It is clear that the contribution of ORMDL3 to the pathogenesis of asthma is quite complex and requires further study.
The presence of maternal allergy is a significant risk factor that increases the likelihood of offspring developing allergy [20]. Pregnancies complicated by asthma can have a significant effect on placental gene expression and potentially contribute to an atopic phenotype in offspring. We have previously shown that pregnancies complicated by maternal asthma are associated with reduced female fetal growth, accompanied by alterations in placental function such as decreased placental 11β-HSD2 [44]. However, the placental immune genes identified that were altered in the presence of maternal asthma including TNFα, IL-1β, IL-6, IL-8, IL10 and IL-5 [13] were not altered in the placentae of children who developed allergy. Furthermore, we did not find any association between maternal asthma and altered gene expression in placentae of children that developed an allergy. This suggests that certain immune mechanisms that are altered in the fetal-placental unit by the presence of maternal asthma are not associated with gene alterations associated with allergy susceptibility in early childhood.
In conclusion, this study has demonstrated a number of placental genes that are altered in association with the development of specific childhood allergic phenotypes and suggests the in utero environment may play a role in programming the immune system. The potential mechanisms driving increased susceptibility to allergy via these altered genes are illustrated in our proposed model in figure 3. Based on this model we propose that abnormal expression of these genes in the placenta may cause a bias towards a Th2 immune profile in the fetal circulation including increased levels of CD4+ cells, increased secretion of anti-inflammatory cytokines and decreased Treg activity as previously indicated by reduced placental FOXP3 expression [10]. As a result, altered programming of fetal immune function may result in increased susceptibility allergic disease.
Acknowledgements
We thank our research midwife Ms Karen Rivers and our asthma research nurse Ms Kate Roberts-Thompson for all their efforts in patient recruitment, sample collection and post-natal follow-ups. We also thank the patients for consenting to take part in our study. This work was supported by the National Health and Medical Research Council Project Grant (APP1002381) and Channel 7 Children's Research Foundation Grant (14850). VLC was supported by a National Health and Medical Research Council Fellowship (APP1041918).
Ethical Statement
This work was approved by the Government of South Australia SA Health Human Research Ethics Committee (TQEH/LMH/MH) and the University of Adelaide Human Research Ethics Committee (2009045).
Author Contributions
ART: study design, coordination, sample collection, experimental work, data analysis, manuscript writing. SME: data analysis, manuscript revision. LEG: data collection, data analysis, manuscript drafting and revision. AO: data collection, sample collection, manuscript revision. ZS: data collection, sample collection, manuscript revision. AT: study design, data collection, study advice and manuscript drafting and revision. SLP: sample collection, data collection and manuscript drafting and revision. MT: study advice and manuscript drafting and revision. RS: study advice and manuscript drafting and revision. VLC: study design, coordination, supervision and manuscript drafting and revision.
Conflict of Interest
The authors do not have any conflicts of interest to declare.
References
-
Hopper JL, Jenkins MA, Carlin JB, Giles GG (1995) Increase in the self-reported prevalence of asthma and hay fever in adults over the last generation: a matched parent-offspring study. Aust J Public Health 19: 120-124.
-
Peat JK, van den Berg RH, Green WF, Mellis CM, Leeder SR, et al. (1994) Changing prevalence of asthma in Australian children. BMJ 308: 1591-1596.
-
AIHW (2005) Asthma in Australia 2005.
-
Prescott SL, Tang ML; Australasian Society of Clinical Immunology and Allergy (2005) The Australasian Society of Clinical Immunology and Allergy position statement: Summary of allergy prevention in children. Med J Aust 182: 464-467.
-
Prescott SL (2003) Early origins of allergic disease: a review of processes and influences during early immune development. Curr Opin Allergy Clin Immunol 3: 125-132.
-
Prescott SL, Clifton V (2009) Asthma and pregnancy: emerging evidence of epigenetic interactions in utero. Curr Opin Allergy Clin Immunol 9: 417-426.
-
Prescott S, Saffery R (2011) The role of epigenetic dysregulation in the epidemic of allergic disease. Clin Epigenetics 2: 223-232.
-
Campbell DE, Boyle RJ, Thornton CA, Prescott SL (2015) Mechanisms of allergic disease - environmental and genetic determinants for the development of allergy. Clin Exp Allergy 45: 844-858.
-
Levy O (2007) Innate immunity of the newborn: basic mechanisms and clinical correlates. Nat Rev Immunol 7: 379-390.
-
Prescott SL, Tulic M, Kumah AO, Richman T, Crook M, et al. (2011) Reduced placental FOXP3 associated with subsequent infant allergic disease. J Allergy Clin Immunol 128: 886-887.
-
Mikaelsson MA, Constancia M, Dent CL, Wilkinson LS, Humby T (2013) Placental programming of anxiety in adulthood revealed by Igf2-null models. Nat Commun 4: 2311.
-
Clifton VL (2010) Review: Sex and the human placenta: mediating differential strategies of fetal growth and survival. Placenta 31 Suppl: S33-39.
-
Scott NM, Hodyl NA, Murphy VE, Osei-Kumah A, Wyper H, et al. (2009) Placental cytokine expression covaries with maternal asthma severity and fetal sex. J Immunol 182: 1411-1420.
-
Scott NM, Hodyl NA, Osei-Kumah A, Stark MJ, Smith R, et al. (2011) The presence of maternal asthma during pregnancy suppresses the placental pro-inflammatory response to an immune challenge in vitro. Placenta 32: 454-461.
-
Worldwide variation in prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and atopic eczema: ISAAC. (1998) The International Study of Asthma and Allergies in Childhood (ISAAC) Steering Committee. Lancet 351: 1225-1232.
-
Asher MI, Keil U, Anderson HR, Beasley R, Crane J, et al. (1995) International Study of Asthma and Allergies in Childhood (ISAAC): rationale and methods. Eur Respir J 8: 483-491.
-
Murphy VE, Clifton VL (2003) Alterations in human placental 11beta-hydroxysteroid dehydrogenase type 1 and 2 with gestational age and labour. Placenta 24: 739-744.
-
Radoniæ A, Thulke S, Mackay IM, Landt O, Siegert W, et al. (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 313: 856-862.
-
Wadzinski TL, Geromini K, McKinley Brewer J, Bansal R, Abdelouahab N, et al. (2014) Endocrine disruption in human placenta: expression of the dioxin-inducible enzyme, cyp1a1, is correlated with that of thyroid hormone-regulated genes. J Clin Endocrinol Metab 99: E2735-2743.
-
Wang QP, Wu KM, Li ZQ, Xue F, Chen W, et al. (2012) Association between maternal allergic rhinitis and asthma on the prevalence of atopic disease in offspring. Int Arch Allergy Immunol 157: 379-386.
-
Khan TK, Palmer DJ, Prescott SL (2015) In-utero exposures and the evolving epidemiology of paediatric allergy. Curr Opin Allergy Clin Immunol 15: 402-408.
-
Camateros P, Tamaoka M, Hassan M, Marino R, Moisan J, et al. (2007) Chronic asthma-induced airway remodeling is prevented by toll-like receptor-7/8 ligand S28463. Am J Respir Crit Care Med 175: 1241-1249.
-
Moisan J, Camateros P, Thuraisingam T, Marion D, Koohsari H, et al. (2006) TLR7 ligand prevents allergen-induced airway hyperresponsiveness and eosinophilia in allergic asthma by a MYD88-dependent and MK2-independent pathway. Am J Physiol Lung Cell Mol Physiol 290: L987-995.
-
Møller-Larsen S, Nyegaard M, Haagerup A, Vestbo J, Kruse TA, et al. (2008) Association analysis identifies TLR7 and TLR8 as novel risk genes in asthma and related disorders. Thorax 63: 1064-1069.
-
Xirakia C, Koltsida O, Stavropoulos A, Thanassopoulou A, Aidinis V, et al. (2010) Toll-like receptor 7-triggered immune response in the lung mediates acute and long-lasting suppression of experimental asthma. Am J Respir Crit Care Med 181: 1207-1216.
-
Sercan O, Stoycheva D, Hammerling GJ, Arnold B, Schuler T (2010) IFN-gamma receptor signaling regulates memory CD8+ T cell differentiation. J Immunol 184: 2855-2862.
-
Bacher N, Raker V, Hofmann C, Graulich E, Schwenk M, et al. (2013) Interferon-α suppresses cAMP to disarm human regulatory T cells. Cancer Res 73: 5647-5656.
-
Moldenhauer LM, Keenihan SN, Hayball JD, Robertson SA (2010) GM-CSF is an essential regulator of T cell activation competence in uterine dendritic cells during early pregnancy in mice. J Immunol 185: 7085-7096.
-
Corry DB, Kiss A, Song LZ, Song L, Xu J, et al. (2004) Overlapping and independent contributions of MMP2 and MMP9 to lung allergic inflammatory cell egression through decreased CC chemokines. FASEB J 18: 995-997.
-
Vermaelen KY, Cataldo D, Tournoy K, Maes T, Dhulst A, et al. (2003) Matrix metalloproteinase-9-mediated dendritic cell recruitment into the airways is a critical step in a mouse model of asthma. J Immunol 171: 1016-1022.
-
Collison A, Siegle JS, Hansbro NG, Kwok CT, Herbert C, et al. (2013) Epigenetic changes associated with disease progression in a mouse model of childhood allergic asthma. Dis Model Mech 6: 993-1000.
-
Krishnamoorthy N, Oriss TB, Paglia M, Fei M, Yarlagadda M, et al. (2008) Activation of c-Kit in dendritic cells regulates T helper cell differentiation and allergic asthma. Nat Med 14: 565-573.
-
Wu W, Wang T, Dong JJ, Liao ZL, Wen FQ (2012) Silencing of c-kit with small interference RNA attenuates inflammation in a murine model of allergic asthma. Int J Mol Med 30: 63-68.
-
Fang Q, Zhao H, Wang A, Gong Y, Liu Q (2011) Association of genetic variants in chromosome 17q21 and adult-onset asthma in a Chinese Han population. BMC Med Genet 12: 133.
-
Galanter J, Choudhry S, Eng C, Nazario S, Rodriguez-Santana JR, et al. (2008) ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med 177: 1194-1200.
-
Hirota T, Harada M, Sakashita M, Doi S, Miyatake A, et al. (2008) Genetic polymorphism regulating ORM1-like 3 (Saccharomyces cerevisiae) expression is associated with childhood atopic asthma in a Japanese population. J Allergy Clin Immunol 121: 769-770.
-
Hrdlickova B, Holla LI (2011) Relationship between the 17q21 locus and adult asthma in a Czech population. Hum Immunol 72: 921-925.
-
Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, et al. (2007) Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448: 470-473.
-
Vercelli D (2008) Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol 8: 169-182.
-
Wan YI, Shrine NR, Soler Artigas M, Wain LV, Blakey JD, et al. (2012) Genome-wide association study to identify genetic determinants of severe asthma. Thorax 67: 762-768.
-
Wu H, Romieu I, Sienra-Monge JJ, Li H, del Rio-Navarro BE, et al. (2009) Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. Allergy 64: 629-635.
-
Ono JG, Worgall TS, Worgall S (2014) 17q21 locus and ORMDL3: an increased risk for childhood asthma. Pediatr Res 75: 165-170.
-
Carreras-Sureda A, Cantero-Recasens G, Rubio-Moscardo F, Kiefer K, Peinelt C, et al. (2013) ORMDL3 modulates store-operated calcium entry and lymphocyte activation. Human molecular genetics 22: 519-530.
-
Murphy VE, Gibson PG, Giles WB, Zakar T, Smith R, et al. (2003) Maternal asthma is associated with reduced female fetal growth. Am J Respir Crit Care Med 168: 1317-1323.
Special issue title: Allergies and Asthma in Children
Handling Editor: Haibin Wang
Assistant Professor
Department of Medicine
Division of Allergy, Inflammation and Immunology
Beth Israel Deaconess Medical Center
Harvard Medical School Boston
USA





