International Journal of Virology and AIDS
Characterization of Human Influenza Viruses Propagated by Madin-Darby Canine Kidney Cells or Embryonated Eggs
Ewan P Plant*, Laura Couzens, Samuel Mindaye, Elizabeth M Coyle, Surender Khurana, Hang Xie, Hana Golding, Maryna C Eichelberger and Zhiping Ye
Division of Viral Products, United States Food and Drug Administration, Maryland 20993, USA
*Corresponding author:
Ewan Plant, United States Food and Drug Administration, 10903 New Hampshire Ave, Silver Spring, Maryland 20993, USA, Tel: +1 240 402-7319, E-mail: ewan.plant@fda.hhs.gov
Int J Virol AIDS, IJVA-2-011, (Volume 2, Issue 2), Research Article; ISSN: 2469-567X
Received: October 14, 2015 | Accepted: November 17, 2015 | Published: November 22, 2015
Citation: Plant EP, Couzens L, Mindaye S, Coyle EM, Khurana S, et al. (2015) Characterization of Human Influenza Viruses Propagated by Madin-Darby Canine Kidney Cells or Embryonated Eggs. Int J Virol AIDS 2:014. 10.23937/2469-567X/1510014
Copyright: © 2015 Plant EP, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
The hemagglutination assay is a commonly used method to titer influenza virus. It relies on the interaction between virus particles and red blood cells being sufficient to prevent the settling of the blood cells. During additional assessment of human influenza virus stocks titrated by the hemagglutination assay we noted that virus grown in cell culture had a greater number of virus particles than analogous virus grown in eggs. Virus grown in both egg and Madin-Darby canine kidney cells are used for vaccine strain selection and to study antigenic variants by serological assays so it is important to know about any differences in the viruses used in the assays.
The goal of this work was to evaluate physical and biological characteristics of virus propagated in either egg allantoic fluid or cell culture. Using paired virus samples, grown in either egg or cell culture, we determined the particle number, hemagglutination titer and plaque forming units for H1N1, H3N2 and B viruses. Neuraminidase activity assays, receptor binding assays and quantification of viral proteins were performed.
Our data confirmed the initial observation of more particles per HA titer when the virus is propagated in cell culture. The cell propagated samples also had a greater proportion of non-infectious particles. Here we discuss possible reasons for the discrepancy between hemagglutination titer and virus particle count and the potential impact on infectivity studies and serological assays.
Keywords
Influenza, Particle count, Hemagglutination, Non-infectious particles
Introduction
Virus titer is a critical variable for many experiments including growth, transmission, infectivity, neutralization and immunology studies. One of the oldest and most widely used methods for titrating influenza viruses is the hemagglutination assay. Hemagglutination by influenza virus was first reported by G.K. Hirst in 1941 [<[ href="#ref1" class="hd">1]. It was noted that influenza virus propagated in eggs could cause chicken red blood cells (RBCs) to link together and this effect could be titrated by sample dilution. Hemagglutination titers were proportional to the titers that were lethal in mice and the method was significantly faster than titration by mouse infection widely used at the time [2]. It was also noted that antiserum from a ferret infected with the same type of influenza could abrogate hemagglutination. This led to the development of the hemagglutination inhibition (HI) assay. The basic HI assay has been described by the World Health Organization (WHO) [3] but because the method is intended for diverse purposes the assay diverges between different laboratories [4]. For example, guinea pig red blood cells (RBCs) have been used to evaluate an H3N2 vaccine [5] and horse RBCs have been used to evaluate an H5N1 vaccine [6]. Agglutination occurs when the virus binds receptors on the RBCs and links them together. The efficiency of this interaction depends on the specificity and affinity of the viral hemagglutinin protein to the receptors on the RBCs [4,7,8]; this interaction could be impacted by the membrane environment, glycosylation of HA, amount of HA per virion, shape of the viral particle, NA activity, interference by HA antibodies and non-specific interference by NA antibodies. Thus, different influenza strains will vary in the number of particles required to agglutinate RBCs which may add a degree of uncertainty to the HI assays. Recent efforts have been made to improve the assay so that a single version can be used for different applications [9]. But additional adjustments may be needed for emerging viruses as the optimal sensitivity of the assay is best achieved by matching the sialic acid binding preference of the virus to cells with an abundance of those receptors [10].
There are many other methods available for quantifying virus including plaque assay, mouse infectious dose, tissue culture infectious dose and egg infectious dose but these are time consuming taking several days to complete. Methods that quantify nucleic acid or protein content (qPCR or HPLC for example) are quicker but require virus specific reagents or additional specificity tests. Counting particles by transmission electron microscopy (TEM) provides good specificity but requires specialized equipment and experienced personnel. Recently several new methods to count particles have been developed including Field-Flow Fractionation-Multiangle Light Scattering [11], Nanoparticle Tracking Analysis [12] and flow cytometry-based approaches like the Virus Counter. Virus Counter quantification of influenza virus has been shown to correlate well with other methods such as qPCR, HPLC and TEM counts [13,14]. An advantage many of these methods have over the hemagglutination assay is that they are not complicated by sialic acid binding preferences.
It has previously been noted that viruses grown in different culture systems have differences in receptor binding specificity [8,15]. In some instances these differences alter the antigenicity of the virus and immunological responses of the host after vaccination [16]. The HI titers of antisera from ferrets infected with either the cell- or egg-grown virus have been shown to be higher against the virus grown in the same culture system ([5] for B/Brisbane/60/2008; [17] for (H3N2) A/Memphis/12/1985). Differences in HI titer are sometimes attributed to changes in the hemagglutinin sequence after viruses are passaged in eggs [8,18] or to changes in the neuraminidase sequence which may contribute to hemagglutination [7]. Supporting the notion that egg-adaptation alters the virus, higher HI titers were observed in an epidemiological study where human sera were tested with cell-propagated virus than with egg-propagated virus [19]. Interestingly, when egg-adapted viruses are further cultured in MDCK cells the virus often retains the egg-like serological characteristics and do not revert back to the cell-like characteristics [20]. Changes to the hemagglutinin by adaptation in eggs have been implicated in low vaccine effectiveness [21]. However it was recently noted that the majority of cell-propagated H3N2 viruses gave lower HI titers compared to egg-propagated viruses [22]. This would suggest that other variables might be involved. The titration of viruses for the HI assay is dependent on the interaction between the virus and the receptors on the RBCs and, as the hemagglutinin has been shown to mutate during adaptation to egg culture [23], we postulated that the amount of virus needed to agglutinate RBCs may depend on the source of the virus. Here we compare the HA titer and particle count of virus propagated in eggs and cell culture and investigate possible causes for observed differences.
Material and Methods
Viruses
The viruses used in this study (Table 1) were provided by WHO Collaborating Centers and National Influenza Centers. Cell and egg isolates of A/Bolivia/559/2013 (H1N1), A/New York/39/2012 (H3N2) and B/Massachusetts/02/2012 were grown on Madin-Darby Canine Kidney (MDCK) cells and in 9-10 day old embryonated eggs respectively.
Virus characterization
All cell derived viruses were grown in cell culture and all egg derived viruses were grown in eggs. Cell propagated virus was grown without serum in Eagles Minimal Essential Media (Catalog #12-125F, Lonza, Allendale, NJ) supplemented with glutamine (Catalog# 35050-061, Thermo Fisher Scientific, Rockford, IL) and TPCK trypsin (Catalog# T3053, Sigma) at 37°C with 5% CO2. At 48 hours post-inoculation culture supernatant or allantoic fluid was harvested, clarified by centrifugation and stored at -80°C. Larger volumes of virus were prepared for sucrose purification by inoculating fifteen eggs with 2.5 x 104 virus particles each or one roller bottle containing approximately 5 x 108 cells in 200 mL culture media with 5 x 105 virus particles. After 48 hours virus was harvested from the culture supernatant or allantoic fluid and pelleted at 30,000 rpm for 90 minutes in a Beckman SW32Ti rotor. The washed pellets were then banded on a 60% sucrose cushion, pelleted and resuspended in 0.5 mL PBS and briefly sonicated to facilitate resuspension. Total protein was determined using a Pierce BCA Protein Assay Kit (Catalog #23225, Thermo Fisher Scientific) according to the manufacturer's instructions.
The hemagglutination assay was performed in 96-well round bottom plates. Duplicate samples of virus (50 μl) were diluted two-fold in PBS using calibrated pipettes. Control wells with no virus were included in every plate. An equal volume of red blood cells (RBCs) was added to all wells and the plates incubated at room temperature for 60 minutes. Hemaggutination titers were recorded as the reciprocal of the dilution of virus in the last well with complete hemagglutination. Freshly prepared turkey RBCs (0.5% in PBS) were used for H1N1 and B virus quantification and freshly prepared guinea pig RBCs (1%) were used for H3N2 virus quantification.
Three independent sucrose purified preparations for each virus were serially diluted and assayed for neuraminidase activity as previously described [24]. Vibrio cholerae neuraminidase (Catalog# N7885, Sigma) was used to generate a standard curve for each assay from which activity was calculated. The receptor binding assay was performed using surface plasmon resonance as previously described [25].
Virus particle counts were determined using a 2100 model Virus Counter (ViroCyt, Boulder, CO) according to the manufacturer's instructions. Virus samples from tissue culture supernatant or from egg allantoic fluid were typically diluted 1:20 in ViroCyt sample dilution buffer before counting. Plaque assays were performed on MDCK cells incubated for 72 hours at 33°C with an EMEM, 0.8% agarose overlay with TPCK trypsin (Catalog# T3053, Sigma).
Virus protein quantification
Samples containing the whole virus were digested and the quantity of individual protein was determined as previously described [26]. Briefly, virus suspension containing 12.5 μg total proteins was taken and appropriate internal standards (0.94 μg rH1 and 0.38 μg rH5) were spiked. The protein mixture was digested following standard procedure of reduction and alkylation. In total, 150-750 ng of protein digests was loaded onto nanoACQUITY UPLC system (Waters) equipped with a reverse phase separation column and the eluting peptides were analyzed online using Xevo G2 MS instrument (Waters Corp). The generated MSE data was processed using Proteinlynx Global Server v.2.4 (PLGS) (Waters Corp) and searched against strain-specific flu viral database (GISAID) using the default search parameters. The label-free protein quantification used MS signal response (the average signal counts of three most intense tryptic peptides per mole of a protein) of the internal standard to estimate the concentration of other detectable proteins in the digestion mixture.
Statistical Analysis
Results were recorded in Microsoft Excel files and the statistical calculations performed using equations included in the software. The minimum number of replicates for each experiment is given in the text.
Results
The ratio of virus particles to hemagglutination titer varies among virus strains
Each year sera from subjects vaccinated with the previous season's influenza vaccine are evaluated for antibodies against currently circulating strains. Egg and cell grown viruses are titrated using the hemagglutination assay to standardize the amount of virus used in serology. To assess the similarity between viruses titrated for serologic assays we performed a particle count on the samples normalized for hemagglutination unit (HAU) (Table 1). The particle counts for these samples ranged from 6.8 x 106 to 2.3 x 108 VP/mL for the H1N1 viruses, 3.8 x 107 to 5.5 x 108 VP/mL for the H3N2 viruses, and 3.7 x 106 to 1.4 x 108 VP/mL for the B viruses. When we compared the particle numbers between the egg and cell propagated samples we observed a trend for more particles from the cell propagated samples (Figure 1). This sampling was from the viruses listed in table 1 and, because a wide range of counts was observed, the difference between the cell and egg propagated samples was not statistically different for the H3N2 and B viruses. A two-tailed unpaired t-test suggested that the difference between the H1N1 cell and egg propagated samples might be significant (p-value = 0.02).
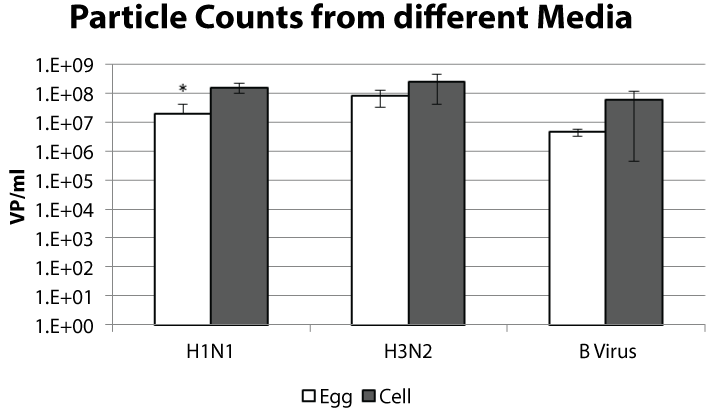
.
Figure 1: The virus samples described in table 1 are graphed by virus type and growth method. The viruses were titrated to a hemagglutination titer of 8 per 50 μl. The average particle count per mL and standard deviation is shown. * p-value < 0.05.
View Figure 1
![]()
Table 1: : HA Normalized Viruses Vary in Particle Number. H1N1 and B viruses were titrated with 0.5% turkey red blood cells and H3N2 viruses were titrated with 1% guinea pig blood cells. Particle counts were performed with virus titrated to a hemagglutination titer of 8 per 50 μl and the VP/HAU calculated.
View Table 1
Cell propagated viruses have lower HA titer per particle
To determine if the observed difference in particle numbers from hemagglutinatin titer normalized egg- and cell-sourced viruses was robust we cultured three paired viruses in egg allantoic fluid or cell culture and looked for a correlation between hemagglutination titer and particle counts. Stocks of A/Bolivia/559/2013 (H1N1), A/New York/39/2012 (H3N2) and B/Massachusetts/02/2012 viruses derived from either cell or egg isolates were prepared. After 48 hours incubation the allantoic fluid or culture supernatant were harvested, purified and virus quantity assessed. We determined virus quantity by three different methods; particle counts, plaque forming units and hemagglutination assay.
The concentrated viruses were serial diluted 1.5:1 in PBS for particle count and hemagglutination assays (Figure 2). Hemagglutination titrations typically use a 2-fold dilution and titers are recorded as such (2, 4, 8, 16...). Here a 1.5:1 dilution factor was used here so that multiple data points within each hemagglutination titer could be captured. In this analysis a clear linear relationship between hemagglutination titer and particle count is observed for all samples with R2 values from 0.84 to 0.95. The robust correlation coefficients (0.96-0.97) for the paired cell and egg propagated virus samples indicate that the difference between the virus pairs was not a function of the dilution. For all three virus pairs there were more virus particles in the cell propagated samples with the same hemagglutination titer as the egg propagated samples. This is in line with the initial observation made with the unpurified viruses used in the hemagglutination assays (Figure 1 and Table 1).
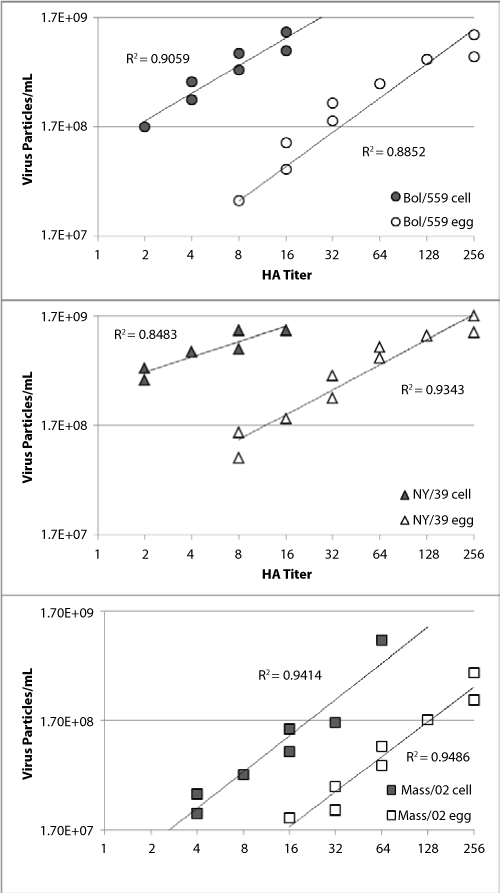
.
Figure 2: HA titers are graphed with the particle count for virus samples propagated in egg (open symbols) or MDCK cells (filled symbols). The paired purified virus samples are A/Bolivia/559/2013 (H1N1; circles), A/New York/39/2012 (H3N2; triangles) and B/Massachusetts/02/2012 (squares).
View Figure 2
Viruses have different infectivity per particle
Infectivity of virus samples is often used as a measure of virus titer, however the infectivity of influenza viruses is not always directly proportional to the number of particles [27,28]. It is possible for virus stocks to lose infectivity, especially with multiple freeze/thaw cycles, indicating that the quality of the viral particle is important [29]. We compared the number of virus particles and plaque forming units as a measure of infectivity from individual eggs or cell culture wells infected with virus. The cell propagated or egg propagated paired virus samples produced similar plaques (Figure 3a). When the particle: plaque ratio was determined for multiple independent virus preparations (n > 7) there was a consistent trend for more non-infectious particles from the cell propagated samples than from the egg propagated samples (Figure 3b). However, two-tailed unpaired t-tests suggested that although this trend was significant for the influenza B virus (p value = 0.04) it was not significant for the influenza A viruses (p values 0.12 and 0.36). There are several variables that could contribute to this effect. First, the timing of the virus harvest might contribute to the number of non-infectious particles. It has been observed in growth curves for some viruses, including influenza, that the number of infective particles increases until a peak is reached and then the number declines [30]. The peak infectious yield for each virus was not determined in this study. Second, the hemagglutinin of cell-propagated viruses may have a different avidity for receptors that could impact the kinetics of infection. Human influenza viruses preferentially bind to α2,6-linked sialic acids [31]. Since receptor binding affinity can also contribute to infectivity we assayed virus binding to α2,6-linked sialic acids using surface plasmon resonance (Figure 3c). We observed substantially more binding by the egg propagated H3N2 virus compared to the cell propagated sample. This trend was not observed for the H1N1 or B viruses. These data suggest that α2,6-linked sialic acid binding alone does not account for the differences in particle number or infectivity between HA normalized cell and egg propagated samples.
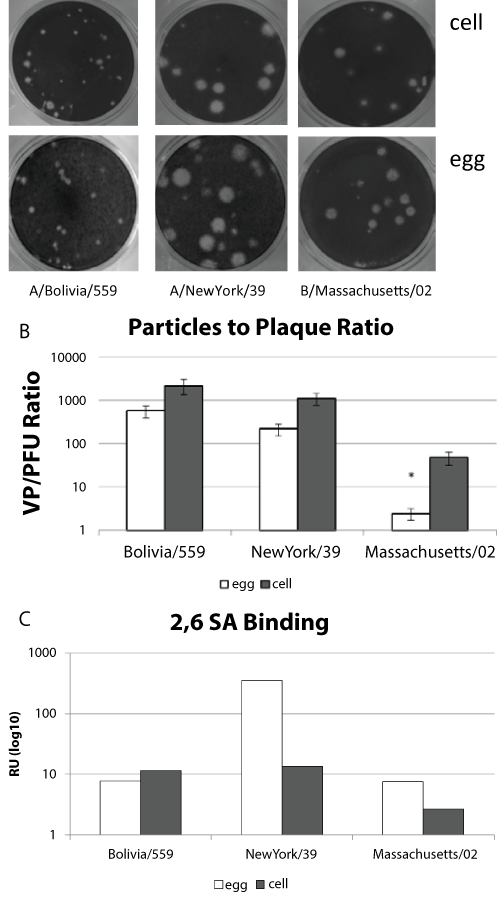
.
Figure 3: Infectious Particles.
(a) Paired virus strains grown in either MDCK cells or egg allantoic fluid were plaqued on MDCK cells.
(b) Independent virus preparations were counted using plaque assay and particle counts. The particle:plaque ratio for each virus strain with standard error is shown.
(c) Relative binding α2, 6-linked sialic acid binding is shown for sucrose purified virus preparations containing 107 virus particles. * p-value < 0.05.
View Figure 3
Virus substrate and neuraminidase activity
Neuraminidase activity plays an important role in virus release. It has been shown that some influenza viruses grown in eggs have higher neuraminidase activity than those grown in cell culture [32]. We determined the neuraminidase activity of viruses propagated in either allantoic fluid or tissue culture. Three independent preparations of each virus from each substrate were assayed. There was higher neuraminidase activity in the egg-derived samples than the cell propagated samples (Figure 4a). An unpaired, two-tail t-test indicated that this might be significant for the H3N2 and B viruses (p values = 0.0004 and 0.01 respectively). Particle counts and HA units were determined for each of the samples and used to normalize the amount of neuraminidase activity (Figure 4b and Figure 4c). Two-tailed unpaired t-tests suggested that the differences between the egg and cell propagated samples were not significant when normalized to particle count (p values = 0.36, 0.56 and 0.22 for the H1N1, H3N2 and B viruses respectively). When the neuraminidase activity was normalized to HAU the t-test suggested that there was no difference in activity between the cell or egg derived viruses (p value = 0.24, 0.07 and 0.72 for the H1N1, H3N2 and B viruses respectively). These data suggest that the amount of neuraminidase activity is similar per virus particle whether it is derived from eggs or cell culture.
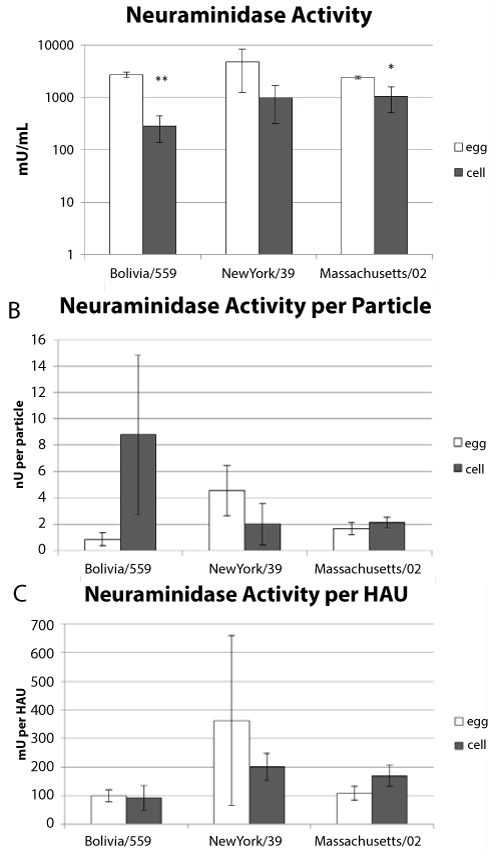
.
Figure 4: Neuraminidase Activity.
(a) Three independent sucrose purified virus samples were assayed for neuraminidase activity for each strain. The average neuraminidase activity and standard deviation is shown.
(b) The neuraminidase activity normalized to particle number with standard error.
(c) The neuraminidase activity normalized to HA titer with standard errors. * p-value < 0.05, ** p-value < 0.01.
View Figure 4
Host cell affects virus protein content
Viral proteins congregate on the host cell membrane before viral budding. The density and dispersion of viral proteins is likely affected by the nature of the host cell membrane. The relative amount of viral proteins present in viruses propagated in either eggs or tissue culture was compared using mass spectrometry (MS). The genome size of each paired virus does not differ, therefore the amount of NP protein per virion should be the same for the egg or cell propagated samples. The amount of hemagglutinin and neuraminidase from the viruses was normalized by expressing it as a ratio with NP (Table 2). In the B/Massachusetts/02 samples the HA:NP ratio was 2-fold higher in the egg preparation when compared to the cell culture preparation (7.7 and 3.6 respectively) indicating more HA is present per egg derived virion. A similar result was observed with the NA:NP ratios (0.7 and 0.28 respectively). Neuraminidase was below the level of quantification for the cell propagated H1N1 and H3N2 samples and was lower in the B/Massachusetts/02 cell grown samples which correlates with the lower neuraminidase activity observed in figure 4a. As the amount of M1 can vary between different viruses and different HA:M1 ratios have been associated with different levels of infectivity [33] the HA:M1 ratios were calculated (Table 2). The HA:M1 ratios were more similar for the egg and cell culture preparations of B/Massachusetts/02 (1.9 and 2.7 respectively) but quite different for the A/Bolivia/559 preparations (0.18 and 5.8 for egg and cell preparations respectively) suggesting the M1 abundance is related to virus type as well as propagation method.
![]()
Table 2: Amount of Viral Protein. Virus proteins detected by mass spectrometry were quantified. The amount of viral protein (femto-moles of HA, NA, NP or M1) detected per nanogram of purified virus loaded is shown. The amount of hemagglutinin and neuraminidase protein is expressed as a ratio with either the NP or M1 proteins. ND: where viral protein was not detected the ratio was not determined.
View Table 2
Discussion
There is evidence that suggests that the propagation method affects virus receptor binding and serological results [18,20]. While H1N1 and B viruses used in the HI assay are titrated with avian RBCs, H3N2 viruses are often titrated with mammalian RBCs because after 1992 some H3N2 viruses were unable to agglutinate chicken RBCs [34]. We have observed that the HA titer measured by avian RBCs of some recent H3N2 virus isolates grown in eggs can be up to 2 fold higher than that titrated by mammalian RBCs whereas there is no difference for cell grown viruses (unpublished data; EP, ZY). This observation correlates well with the increased α2,6-linked sialic acid binding observed for the A/New York/39/2012 egg grown virus used in this work. However, a similar change in sialic acid binding was not observed for the H1N1 and B viruses used in this work indicating that the novel finding of greater particle numbers in cell propagated viruses is not due solely to egg adaptation and sialic acid binding.
Differences in the amount of viral protein detected by mass spectrometry for each strain grown using either eggs or cell culture indicate that influenza A and B viruses grow quite differently from each other. Harvesting virus after 48 hour resulted in a much lower amount of influenza a virus from the cell grown cultures but this was not the case for the influenza B strain used here. Interestingly, while there was greater HA content in the cell grown influenza B samples compared to the egg grown, the ratios of HA:NP and NA:NP were lower suggesting each cell grown particle has less HA and NA on its surface. This lower proportion of HA per virus particle correlates well with the increased particle counts observed for cell grown samples compared to egg grown samples with the same hemagglutination titer. While influenza virus particles exhibit two major morphologies, spherical and filamentous, there is significant structural variability in viruses from a particular strain [35,36,]. Different PR/8 strains used to create reassortant viruses also yield different proportions of spherical and filamentous particles [37]. We hypothesize that this variation is also skewed when viruses are propagated using different growth methods. This was exemplified by the cell-derived H1N1 virus that had a much higher HA:M1 ratio than egg grown virus.
The total number of virus particles required to agglutinate the RBCs will vary from virus to virus depending on factors such as the density of hemagglutinin on the surface of the virus and the avidity for the receptors [34,38]. In our hands there was significant variation in the H1N1, H3N2 and B virus particle numbers that were required to reach an HA titer of 8 per 50 μl. Theoretically this difference in virus particle number could affect the HI assay results because the number of antibodies required to sequester different virus types away from the RBCs to prevent agglutination is likely to be proportional to the amount of HA on each virus particle. However, lower HI titers have only been observed for cell grown H3N2 viruses. It has been suggested that amino acid substitutions in the hemagglutinin at positions 186 and 194 were involved. It has previously been noted that substitutions at those positions may be due to egg adaptation and alter the antigenicity of the virus [23,39]. In recent WHO and Vaccine and Related Biological Products Advisory Committee meetings it has been noted that the inhibition of some cell grown H3N2 viruses has been less than the inhibition of egg grown viruses (http://www.who.int/influenza/vaccines/virus/recommendations/201302_recommendation.pdf?ua=1, http://www.fda.gov/downloads/AdvisoryCommittees/CommitteesMeetingMaterials/BloodVaccinesandOtherBiologics/VaccinesandRelatedBiologicalProductsAdvisoryCommittee/UCM389690.pdf, accessed August 3, 2015). This trend has been observed for paired virus samples, that is, the same virus isolated and cultivated in different media. This data was obtained with human sera from subjects vaccinated with an egg based vaccine and the same trend is observed with ferret sera. While some differences may be attributed to the virus adapting to the eggs the genomic sequence differences don't always map to sites known to affect receptor binding. For example, we observed no change in α2,6-linked sialic acid binding for cell and egg propagated H1N1 and B virus even though there was a 10-fold difference in particle number.
Other assays that rely on measures of infectivity might be affected by the method of propagation. Our data indicated that cell propagated virus with the same infectious titer as egg propagated virus have more non-infectious particles. It is not clear if these particles interact with other molecules, such as receptors and antibodies, in the same way as the infectious particles. Large numbers of non-infectious particles could sequester antibodies during challenge experiments or microneutralization assays. The presence of non-infectious particles in challenge material could also result in the production of antibodies that inadequately clear infective virus. Our data indicate that, except for the H3N2 virus, the method of propagation did not affect receptor binding yet there was still a difference in the number of particles per hemagglutination unit. This suggests that other factors such as the virus size or morphology play a significant role in virus interactions. Differences in particle size might affect the virus transmission or the stability of viruses in the environment. Further work is required to identify and quantify the roles these factors may have.
Our data demonstrate that the culture medium affects the hemagglutinating ability of virus particles generated. A greater number of particles per hemagglutination unit were observed when influenza virus was cultured in MDCK cells as compared to eggs. The increase in particle number was more easily observed with sucrose purified samples suggesting that factors derived from the culture medium may also affect hemagglutination. Although a difference in hemagglutination inhibition has been reported between egg- and cell-propagated H3N2 viruses at WHO and VRBPAC meetings it has not been reported for H1N1 or B viruses. Our sialic acid binding results are in line with other studies that indicate differences observed for H3N2 viruses may be due to increased binding after egg adaptation. Cell propagation also resulted in an increase in the number of particles per infectious unit under the similar growth and harvest conditions. The difference in the proportion of infectious particles may be related to differences in HA content per virion, however, factors such as overall virus charge and aggregation of particles may also contribute to avidity between the hemagglutinin and receptors. Further studies will establish whether the HI assay can be modified to counteract low hemagglutination titers measured in assays using cell propagated H3N2 viruses.
Acknowledgments
We would like to thank ViroCyt personnel for providing technical assistance with the Virus Counter. This work was supported in part by the Biomedical Advanced Research and Development Authority, Department of Health and Human Services.
References
-
Hirst GK (1941) The agglutination of red cells by allantoic fluid of chick embryos infected with influenza virus. Science 94: 22-23.
-
Hirst GK (1942) The quantitative determination of influenza virus and antibodies by means of red cell agglutination. J Exp Med 75: 49-64.
-
World Health Organization.Global Influenza Surveillance Network (2011) Manual for the laboratory diagnosis and virological surveillance of influenza.
-
Wood JM, Major D, Heath A, Newman RW, Hoschler K, et al. (2012) Reproducibility of serology assays for pandemic influenza H1N1: collaborative study to evaluate a candidate WHO International Standard. Vaccine 30: 210-217.
-
Kishida N, Fujisaki S, Yokoyama M, Sato H, Saito R, et al. (2012) Evaluation of influenza virus A/H3N2 and B vaccines on the basis of cross-reactivity of postvaccination human serum antibodies against influenza viruses A/H3N2 and B isolated in MDCK cells and embryonated hen eggs. Clin Vaccine Immunol 19: 897-908.
-
Noah DL, Hill H, Hines D, White EL, Wolff MC (2009) Qualification of the hemagglutination inhibition assay in support of pandemic influenza vaccine licensure. Clin Vaccine Immunol 16: 558-566.
-
Lin YP, Gregory V, Collins P, Kloess J, Wharton S, et al. (2010) Neuraminidase receptor binding variants of human influenza A(H3N2) viruses resulting from substitution of aspartic acid 151 in the catalytic site: a role in virus attachment? J Virol 84: 6769-6781.
-
Stevens J, Chen LM, Carney PJ, Garten R, Foust A, et al. (2010) Receptor specificity of influenza A H3N2 viruses isolated in mammalian cells and embryonated chicken eggs. J Virol 84: 8287-8299.
-
Morokutti A, Redlberger-Fritz M, Nakowitsch S, Krenn BM, Wressnigg N, et al. (2013) Validation of the modified hemagglutination inhibition assay (mHAI), a robust and sensitive serological test for analysis of influenza virus-specific immune response. J Clin Virol 56: 323-330.
-
Pawar SD, Parkhi SS, Koratkar SS, Mishra AC (2012) Receptor specificity and erythrocyte binding preferences of avian influenza viruses isolated from India. Virol J 9: 251.
-
Bousse T, Shore DA, Goldsmith CS, Hossain MJ, Jang Y, et al. (2013) Quantitation of influenza virus using field flow fractionation and multi-angle light scattering for quantifying influenza A particles. J Virol Methods 193: 589-596.
-
Kramberger P, Ciringer M, A trancar A, Peterka M (2012) Evaluation of nanoparticle tracking analysis for total virus particle determination. Virol J 9: 265.
-
Stepp PC, Ranno KA, Dawson ED, Rowlen KL, Ferris MM (2011) Comparing H1N1 Virus Quantification with a Unique Flow Cytometer and Quantitative PCR. Bio Process International 9: 50-56.
-
Transfiguracion J, Manceur AP, Petiot E, Thompson CM, Kamen AA (2015) Particle quantification of influenza viruses by high performance liquid chromatography. Vaccine 33: 78-84.
-
Schild GC, Oxford JS, de Jong JC, Webster RG (1983) Evidence for host-cell selection of influenza virus antigenic variants. Nature 303: 706-709.
-
Alymova IV, Kodihalli S, Govorkova EA, Fanget B, Gerdil C, et al. (1998) Immunogenicity and protective efficacy in mice of influenza B virus vaccines grown in mammalian cells or embryonated chicken eggs. J Virol 72: 4472-4477.
-
Katz JM, Webster RG (1989) Efficacy of inactivated influenza A virus (H3N2) vaccines grown in mammalian cells or embryonated eggs. J Infect Dis 160: 191-198.
-
Robertson JS, Naeve CW, Webster RG, Bootman JS, Newman R, et al. (1985) Alterations in the hemagglutinin associated with adaptation of influenza B virus to growth in eggs. Virology 143: 166-174.
-
Pyhala R, Pyhala L, Valle M, Aho K (1987) Egg-grown and tissue-culture-grown variants of influenza A (H3N2) virus with special attention to their use as antigens in seroepidemiology. Epidemiol Infect 99: 745-753.
-
Oxford JS, Corcoran T, Knott R, Bates J, Bartolomei O, et al. (1987) Serological studies with influenza A (H1N1) viruses cultivated in eggs or in a canine kidney cell line (MDCK). Bull World Health Organ 65: 181-187.
-
Skowronski DM, Janjua NZ, De Serres G, Sabaiduc S, Eshaghi A, et al. (2014) Low 2012-13 influenza vaccine effectiveness associated with mutation in the egg-adapted H3N2 vaccine strain not antigenic drift in circulating viruses. PLoS One. 9: e92153.
-
Barr IG, Russell C, Besselaar TG, Cox NJ, Daniels RS, et al. (2014) Writing Committee of the World Health Organization Consultation on Northern Hemisphere Influenza Vaccine Composition for 2013-2014. WHO recommendations for the viruses used in the 2013-2014 Northern Hemisphere influenza vaccine: Epidemiology, antigenic and genetic characteristics of influenza A(H1N1)pdm09, A(H3N2) and B influenza viruses collected from October 2012 to January 2013. Vaccine. 32: 4713-4725.
-
Chen Z, Zhou H, Jin H (2010) The impact of key amino acid substitutions in the hemagglutinin of influenza A (H3N2) viruses on vaccine production and antibody response. Vaccine 28: 4079-4085.
-
Sultana I, Gao J, Markoff L, Eichelberger MC (2011) Influenza neuraminidase-inhibiting antibodies are induced in the presence of zanamivir. Vaccine 29: 2601-2606.
-
Khurana S, Verma S, Verma N, Crevar CJ, Carter DM, et al. (2010) Properly folded bacterially expressed H1N1 hemagglutinin globular head and ectodomain vaccines protect ferrets against H1N1 pandemic influenza virus. PLoS One 5: e11548.
-
Getie-Kebtie M, Sultana I, Eichelberger M, Alterman M (2013) Label-free mass spectrometry-based quantification of hemagglutinin and neuraminidase in influenza virus preparations and vaccines. Influenza Other Respir Viruses 7: 521-530.
-
Brooke CB, Ince WL, Wrammert J, Ahmed R, Wilson PC, et al. (2013) Most influenza a virions fail to express at least one essential viral protein. J Virol 87: 3155-3162.
-
Frensing T, Heldt FS, Pflugmacher A, Behrendt I, Jordan I, et al. (2013) Continuous influenza virus production in cell culture shows a periodic accumulation of defective interfering particles. PLoS One 8: e72288.
-
Tyrrell Da, Horsfall F Jr (1954) Disruption of influenza virus; properties of degradation products of the virus particle. J Exp Med 99: 321-342.
-
Smith AM, Adler FR, McAuley JL, Gutenkunst RN, Ribeiro RM, et al. (2011) Effect of 1918 PB1-F2 expression on influenza A virus infection kinetics. PLoS Comput Biol 7: e1001081.
-
Thompson CI, Barclay WS, Zambon MC, Pickles RJ (2006) Infection of human airway epithelium by human and avian strains of influenza a virus. J Virol 80: 8060-8068.
-
Govorkova EA, Kodihalli S, Alymova IV, Fanget B, Webster RG (1999) Growth and immunogenicity of influenza viruses cultivated in Vero or MDCK cells and in embryonated chicken eggs. Dev Biol Stand 98: 39-51.
-
Ruigrok RW, Krijgsman PC, de Ronde-Verloop FM, de Jong JC (1985) Natural heterogeneity of shape, infectivity and protein composition in an influenza A (H3N2) virus preparation. Virus Res 3: 69-76.
-
Kumari K, Gulati S, Smith DF, Gulati U, Cummings RD, et al. (2007) Receptor binding specificity of recent human H3N2 influenza viruses. Virol J 4: 42.
-
Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, et al. (2006) Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci USA 103: 19123-19127.
-
Katz G, Benkarroum Y, Wei H, Rice WJ, Bucher D, et al. (2014) Morphology of influenza B/Lee/40 determined by cryo-electron microscopy. PLoS One 9: e88288.
-
Johnson A, Chen LM, Winne E, Santana W, Metcalfe MG, et al. (2015) Identification of Influenza A/PR/8/34 Donor Viruses Imparting High Hemagglutinin Yields to Candidate Vaccine Viruses in Eggs. PLoS One 10: e0128982.
-
Wasilewski S, Calder LJ, Grant T, Rosenthal PB (2012) Distribution of surface glycoproteins on influenza a virus determined by electron cryotomography. Vaccine 30: 7368-7373.
-
Hartgroves LC, Koudstaal W, McLeod C, Moncorge O, Thompson CI, et al. (2010) Rapid generation of a well-matched vaccine seed from a modern influenza A virus primary isolate without recourse to eggs. Vaccine 28: 2973-2979.





