Journal of Genetics and Genome Research
Sequence Characterization of Bovine Antisense to Insulin-Like Growth Factor Type 2 Receptor Non-Coding RNA (AIRN)
W T Farmer, J R Sommer and C E Farin*
Department of Animal Science, North Carolina State University, Raleigh, United States
*Corresponding author:
C E Farin, Department of Animal Science, North Carolina State University, Raleigh, NC 27695, United States, E-mail: char_farin@ncsu.edu
J Genet Genome Res, JGGR-3-026, (Volume 3, Issue 2), Review Article; ISSN: 2378-3648
Received: May 26, 2016 | Accepted: August 18, 2016 | Published: August 22, 2016
Citation: Farmer WT, Sommer JR, Farin CE (2016) Sequence Characterization of Bovine Antisense to Insulin-Like Growth Factor Type 2 Receptor Non-Coding RNA (AIRN). J Genet Genome Res 3:026. 10.23937/2378-3648/1410026
Copyright: © 2016 Farmer WT, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Bovine insulin-like growth factor type 2 receptor (IGF2R) is an imprinted gene whose aberrant expression has been implicated in development of abnormal offspring syndrome. Bovine AIRN (AIRN) is expressed in post-implantation fetal tissues coinciding with imprinted expression of IGF2R. Although expression patterns of bovine AIRN have been reported based on PCR analysis, characteristics of this transcript are unknown. Therefore, the objective of this work was to sequence characterize the AIRN ncRNA transcript. PCR primer sets (n = 19) were designed based on genomic sequence to “walk” down the predicted AIRN ncRNA sequence. Total RNA extracted from gestational Day 150 bovine fetal liver was used as source material for analysis. Extracted RNA was DNase-treated prior to cDNA synthesis, PCR amplified, and sequenced. A putative bAIRN promoter was located 623 base-pairs upstream of differentially methylated region 2 (DMR2) within intron 2 of IGF2R. A polyadenylation signal was found 117 kb downstream of the promoter. Primer sets designed upstream of the promoter as well as downstream of the polyadenylation signal yielded no PCR amplicons, suggesting that the length of AIRN is approximately 117 kb. In conclusion, bovine AIRN appears to be an antisense transcript of approximately 117 kb in length with a promoter region located 623 bp upstream of DMR2 within intron 2 of IGF2R.
Keywords
AIRN, Antisense RNA, Bovine, IGF2R, Non-coding RNA, Sequence
Introduction
Transfer of in vitro produced (IVP) or somatic cell nuclear transfer (SCNT) manipulated bovine embryos results in a proportion of conceptuses, fetuses, and offspring that exhibit developmental abnormalities collectively referred to as Abnormal Offspring Syndrome (AOS) [1]. Live offspring are generated from 45% of transferred IVP embryos with 5% to 20% of those exhibiting abnormalities depending on the culture system used [1]. Some of the abnormalities are hypothesized to be derived from the inadequacy of the in vitro culture environment and disruption of epigenetic patterns regulating parent-specific expression of imprinted genes [2].
The insulin-like growth factor type 2 receptor (IGF2R) is an imprinted gene whose aberrant expression has been directly related to the overgrowth phenotype following the transfer of IVP embryos in sheep and implicated in overgrowth of bovine IVP fetuses [3-6]. However, little is known about how imprinted expression of IGF2R is regulated in cattle. The maternal bovine IGF2R allele is preferentially expressed and the paternal IGF2R allele is repressed in post-implantation tissues [7]. Silencing of paternal IGF2R coincides with AIRN expression and acquisition of DNA methylation at the paternal IGF2R promoter. Recently, it has been demonstrated that the mechanisms governing imprinted expression of IGF2R may be tissue specific since the degree to which paternal IGF2R is repressed differs between endodermal, mesodermal and ectodermal origins [8]. Because the sequence characteristics of bovine AIRN are not known, the objective of the present study was to characterize the bovine AIRN non-coding RNA sequence.
Material and Methods
Production of Day 150 Fetuses
All procedures and protocols involving the use of animals in this study were approved by the Institutional Animal Care and Use Committee at North Carolina State University. In vivo embryos were produced using superovulated Holstein cows as embryo donors. Cows received two intramuscular (i.m.) injections of 25 mg Prostaglandin F2α (PGF; Lutalyse; Pfizer Animal Health, USA) administered 14 days apart to synchronize estrus. Between Days 10 and 13 of the estrous cycle (Day 0 = estrus), cows received i.m. injections of 20 to 32 mg follicle stimulating hormone (FSH; FSH-P; Schering-Plough, Piscataway, NJ) in decreasing doses over a 4-day period. Estrus was induced by i.m. injection of 25 mg of PGF2α on the morning and evening of the third day of FSH treatment. Cows were artificially inseminated 12 to 24 hours after the first observed standing estrus with frozen thawed semen from a proven Holstein bull. Embryos were collected non-surgically on Day 7 (Day 0 = first detected estrus) by uterine lavage and evaluated for stage of development and grade [9].
Cross-bred Angus heifers were given two i.m. injections of 25 mg PGF 10 to 12 days apart to synchronize estrus and were used as embryo recipients. On Day 7 of the estrous cycle (Day 0 = estrus), a single Grade 1 in vivo produced blastocyst was transferred non-surgically into the uterus and at Day 150 of gestation, heifers were slaughtered and fetuses recovered. Fetal liver samples were collected and immediately snap frozen in liquid nitrogen and stored at -80°C for extraction of whole cell RNA (wcRNA).
RNA extraction of Day 150 bovine fetal liver
Whole cell (wcRNA) from a gestational Day 150 bovine fetal liver was extracted as previously described [10]. Briefly, frozen tissue (25 to 40 mg) was removed from -80°C storage, weighed, placed in a mortar, covered with liquid nitrogen, and subsequently crushed to a fine powder. The powder was homogenized (Brinkmann Homogenizer PT 10/35; Westbury, NY) and dissociated in lysis solution with mercapto-ethanol. RNA was extracted using a total RNA extraction kit (Sigma, GenElute Total Mammalian RNA Extraction Kit) following the manufacturer’s protocols. The quality and integrity of the wcRNA was assessed by nanodrop using ratio of absorbances at 260 nm and 280 nm, which yielded an A260/A280 ratio of 2.11 for gestational Day 150 bovine fetal liver.
cDNA synthesis
Prior to cDNA synthesis, 2 μg of wcRNA from a previously extracted fetal liver sample was DNase treated by incubation of the wcRNA with 1.5 μl of DNase and 2 μl of DNase buffer at 37°C for 20 minutes. Following the manufacturer’s instructions, 2 μg of DNase-treated wcRNA was incubated with 1 μg of random primers (Promega; Madison, WI), 1 μl of 10 mM dNTP mix (PCR Nucleotide Mix, Roche; Mannheim, Germany) and distilled water at 65°C for 5 minutes. After placement on ice for 1 minute, samples were incubated with 4 μl of 5X First Strand Buffer, 1 μl of 0.1 M DTT, and 1 μl of reverse transcriptase (200 U/μl); (Superscript III, Invitrogen; Carlsbad, CA) at 25°C for 5 minutes. This was followed by incubation at 50°C for 60 minutes and inactivation by heating to 70°C for 15 minutes. The synthesized cDNA was subjected to purification using the QIAquick Purification Kit (Qiagen; Qiagen Sciences, MD) according to the manufacturer’s instructions.
Bovine AIRN primers
Bovine AIRN primers were designed using the known bovine genomic DNA sequence of chromosome 9 UMD 3.1 Primary Assembly AC_000166.1 (97,540,000 bp to 97,710,000 bp). All AIRN primer sets (Table 1) were designed using the primer design program, Primer3Plus (Untegrasser and Nijveen 2007, Wageninger University). All amplification products were sequence verified.
![]()
Table 1: Primers used for RT-PCR to detect the non-coding RNA, AIRN.
View Table 1
Reverse transcription-polymerase chain reaction (RT-PCR)
PCR reactions consisted of a 20 μl reaction volume that contained 100 ng of cDNA, 10 mM dNTP mix, Taq DNA polymerase (1.25 U per 20 μl reaction), sense and anti-sense primers (20 ng of each) and PCR water. PCR reactions were performed in a PTC-100 thermocycler (MJ Research, Inc., Watertown, MA). Annealing temperatures varied (Table 1) and were specific for each primer set evaluated in the primer walk.
Results and Discussion
In cattle, AIRN ncRNA is transcribed from the paternal allele in an antisense direction from a transcriptional start site located in exon 2 of IGF2R continuing into intron 1 of the neighboring protein-coding gene, MAS1 (Figure 1A). Within intron 2 lies a differentially methylated region, DMR2 (Figure 1B), that is 2620 bps in length and is unmethylated on the paternal allele and methylated on the maternal one [5]. Moving in an antisense direction, a putative AIRN transcriptional start site was identified 623 bp upstream of the DMR. The promoter region was identified by analyzing the genomic DNA sequence (AC_000166, 97664989 to 97664875) upstream of the DMR for essential promoter elements. A TATA-box was identified 20 bp upstream of the putative transcriptional start site [11,12] (Figure 2). An Activator Protein-1 (AP1) binding site, also known as a 12-O-Tetradecanoylphorbol-13-acetate (TPA) response element, was located upstream of the TATA-box [13]. In addition, downstream of the TATA-box we found an initiation response element (INR) and a down-stream promoter element.
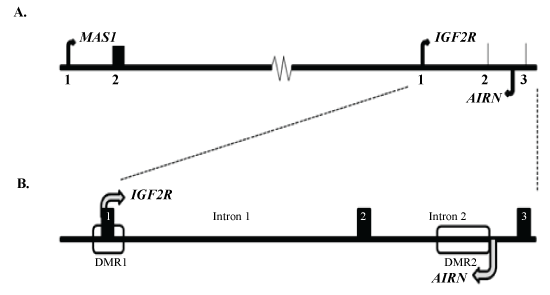
Figure 1: Illustration of the bovine gene region on Chromosome 9 specific to IGF2R and AIRN.
A. Region illustrating the orientation of the sense genes MAS1 and IGF2R to the antisense gene for the non-coding RNA, AIRN. Numbers indicate exons 1 and 2 or exons 1, 2, and 3 of the MAS1 and IGF2R gene sequences, respectively; B. Illustration of the overlap between IGF2R and AIRN. The numbered blocks represent exons 1, 2, and 3 of the IGF2R gene sequence. The open rectangles indicate the location of Differentially Methylated Regions 1 and 2 (DMR1 and DMR2) with the IGF2R gene sequence.
View Figure 1
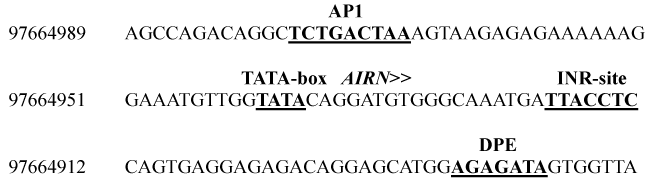
Figure 2: Putative promoter for AIRN. Sequence showing transcription initiation site and consensus binding sites of core promoter elements. Nucleotide numbers refer to the location on chromosome 9 Primary Assembly UMD 3.1 (ACC_000166).
View Figure 2
Interestingly, as has been observed in the murine Airn sequence, there are 4 initiation sites that are clustered together in a 200 bp region in the bovine AIRN promoter region [11]. Also intriguing, is the observation that spread out over a 500 bp region are various promoter elements. Within this region are additional TATA boxes, CAAT boxes, AP1 binding sites, INR elements, and DPEs. These observations are consistent with those made of the murine promoter for Airn [11].
Primers for AIRN (Sets 1 and 2) were designed to amplify regions upstream of the putative promoter and downstream of IGF2R exon 3 (Figure 3A). These upstream primers were subjected to varying PCR conditions, including varied concentrations of cDNA template (50 ng, 100 ng, 150 ng), varied annealing temperatures (50 to 65°C), and altered annealing and extension times (10 to 60s). All attempts to generate a PCR amplicon within this region failed. This suggests that the true promoter region for bovine AIRN is located approximately 623 bp upstream of DMR2 within intron 2 of IGF2R.
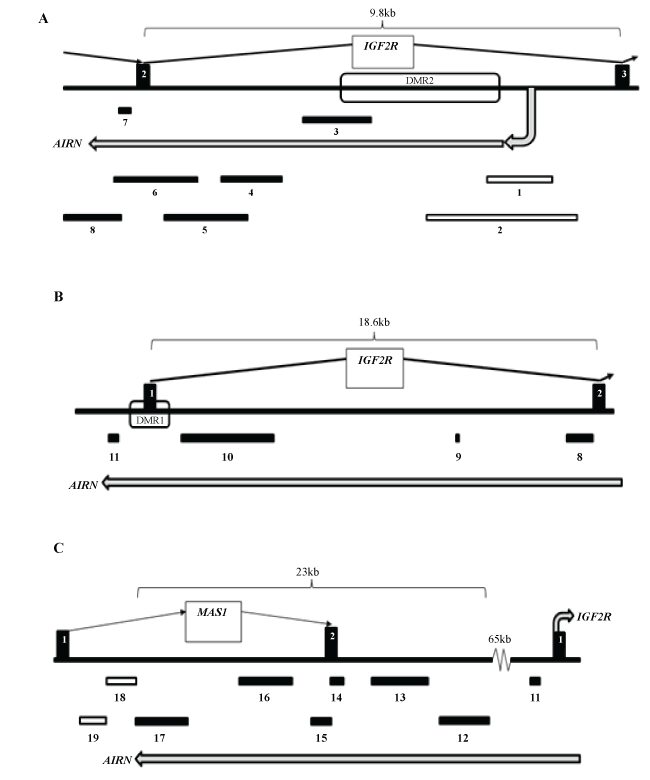
Figure 3: (A) Illustration of the overlap between IGF2R and AIRN at exons 2 and 3 of IGF2R. Vertical Bars depict exons 2 and 3 of the IGF2R gene sequence connected by arrows indicating direction of transcription. Horizontal Bars 1 to 8 indicate the location of PCR primer products for AIRN through the first 10 kb of transcription; (B) Illustration of the overlap between IGF2R and AIRN between exons 1 and 2 of IGF2R. Vertical bars depict Exons 1 and 2 of the IGF2R gene sequence connected by arrows indicating the direction of transcription. Horizontal Bars 8 to 11 indicate the location of PCR primer products for AIRN from 10 kb to 29 kb of the putative AIRN transcript; (C) Illustration of the overlap between MAS1 and AIRN and depiction of PCR amplicons for AIRN for the terminal portion of transcription. Vertical Bars depict the exons 1 and 2 or exon 1 of the MAS1 and IGF2R gene sequences, respectively. Horizontal Bars 12 to 19 indicate the location of PCR primer products for AIRN through the last 23 kb of AIRN transcription. For all panels; (A-C) open horizontal bars indicate that amplicons were not generated, closed horizontal bars indicate that PCR amplicons for AIRN were produced.
View Figure 3
A series of primers for AIRN (n = 19) were designed to PCR amplify regions throughout the predicted length of bAIRN (Figure 3). The PCR amplicons generated covered a total of 27,202 bps of the predicted AIRN transcript. Representative gel images of amplicons produced from PCR primer sets are illustrated in Figure 4.
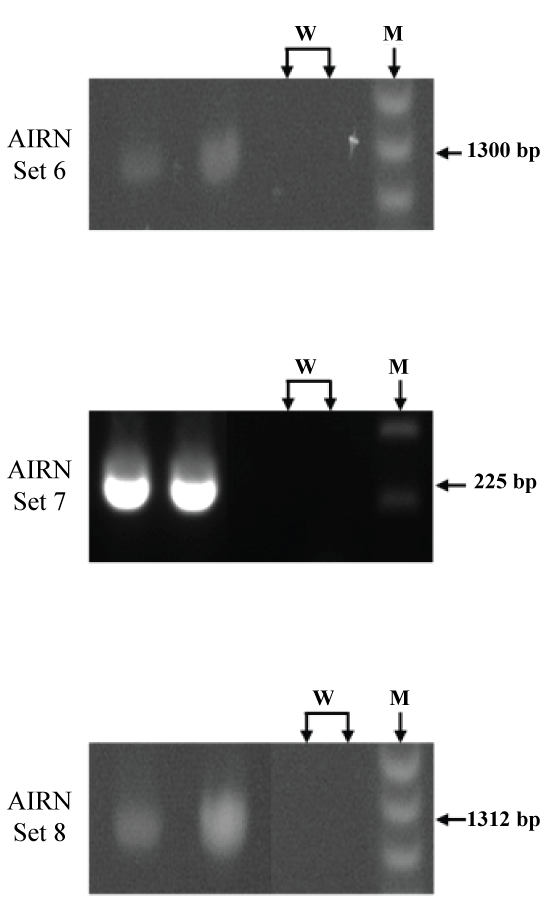
Figure 4: Representative gel images of PCR amplicons for bovine AIRN non-coding RNA. Primer sets 6, 7, and 8 (W, Water Blank; M, Ladder Marker).
View Figure 4
A putative poly-A signal (AATAAA) was identified 270 bps downstream of AIRN primer set 17 (Figure 5). Two additional AIRN primer sets (18 and 19) were designed to amplify products downstream of the poly-A signal (Figure 3C). Amplicons were not obtained from PCR amplification using these primer sets. Therefore, the putative length of bovine AIRN is 117 kb. Amplification of PCR products from primers spread over a 120 kb region from a putative promoter in intron 2 of IGF2R to a putative poly-A signal in intron 1 of MAS1 suggest that AIRN is intron-less. However, because only 23% of the putative AIRN noncoding RNA was sequenced in this effort, the possibility remains that the bovine AIRN transcript may be shorter than 117 kb or may contain introns. Additional poly-A signals (n = 3) can be found throughout the region between the second exon of MAS1 and the first exon of IGF2R. These additional poly-A signals may represent additional transcript termination sites.
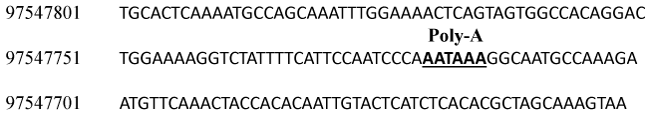
Figure 5: Putative polyadenylation (Poly-A) signal for bovine AIRN. Nucleotide numbers refer to the location on chromosome 9 Primary Assembly UMD 3.1 (ACC_000166).
View Figure 5
Figure 6: Supplemental Material: Individual gel images of PCR amplicons for bovine AIRN non-coding RNA. Set numbers correspond to PCR primer sets listed in table 1 (W, Water Blank; M, Ladder Marker).
View Figure 6
The murine Airn ncRNA sequence has been reported and was found to be 118 kb in length [11,14]. Although the length of bovine AIRN ncRNA appears to be similar to that of the mouse, there is a lack of sequence homology between the two species and some of the core promoter elements also differ. Eukaryotic genomes exhibit a vast range of ncRNAs, however, closer examination of these lncRNAs demonstrated that they may not share close sequence homology between mammalian species [15]. Differences in sequence homology may arise from the presence of transposable elements that are common to mature lncRNAs but rarely occur in transcripts from protein-coding genes [15-17]. Therefore, it may not be surprising that bovine AIRN and murine Airn do not share extensive sequence homology throughout their given lengths.
Conclusion
Bovine AIRN has been characterized as a long non-coding RNA that is estimated to be approximately 117 kb in length. The transcriptional start site is located outside of the imprint control region located within intron 2 of IGF2R. Transcription appears to be continuous from the putative promoter to a poly-A site within intron 1 of MAS1. The AIRN transcript is repeat rich and collinear with the genomic sequence. Based on the known function of murine Airn [18,19], it is presumed that bovine AIRN functions to regulate imprinted expression of IGF2R [20,21]. Further studies will be needed to confirm the functional role of bovine AIRN in regulating imprinted expression of IGF2R or other protein-coding genes.
References
-
Farin PW, Piedrahita JA, Farin CE (2006) Errors in development of fetuses and placentas from in vitro-produced bovine embryos. Theriogenology 65: 178-191.
-
Wrenzycki C, Herrmann D, Lucas-Hahn A, Lemme E, Korsawe K, et al. (2004) Gene expression patterns in in vitro-produced and somatic nuclear transfer-derived preimplantation bovine embryos: relationship to the large offspring syndrome? Anim Reprod Sci 82-83: 593-603.
-
Farin CE, Alexander JE, Farin PW (2010) Expression of messenger RNAs for insulin-like growth factors and their receptors in bovine fetuses at early gestation from embryos produced in vivo or in vitro. Theriogenology 74: 1288-1295.
-
Li S, Yanxin Li, Shuyang Yu, Weihua Du, Lei Zhang, et al. (2007) Expression of insulin-like growth factors systems in cloned cattle dead within hours after birth. Mol Reprod Dev 74: 397-402.
-
Long, JE, X Cai (2007) Igf-2r expression regulated by epigenetic modification and the locus of gene imprinting disrupted in cloned cattle. Gene 388: 125-134.
-
Young LE, Fernandes K, McEvoy TG, Butterwith SC, Gutierrez CG, et al. (2001) Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat Genet 27: 153-154.
-
Suteevun-Phermthai T, Curchoe CL, Evans AC, Boland E, Rizos D, et al. (2009) Allelic switching of the imprinted IGF2R gene in cloned bovine fetuses and calves. Anim Reprod Sci 116: 19-27.
-
Daniela Bebbere, Stefan Bauersachs, Rainer W Fürst, Horst-Dieter Reichenbach, Myriam Reichenbach, et al. (2013) Tissue-specific and minor inter-individual variation in imprinting of IGF2R is a common feature of Bos taurus Concepti and not correlated with fetal weight. PLoS One 8: e59564.
-
(2005) IETS. International Embryo Transfer Society Manual. International Embryo Transfer Society.
-
Blondin P, Farin PW, Crosier AE, Alexander JE, Farin C E (2000) In vitro production of embryos alters levels of insulin-like growth factor-II messenger ribonucleic acid in bovine fetuses 63 days after transfer. Biol Reprod 62: 384-389.
-
Lyle R, Watanabe D, te Vruchte D, Lerchner W, Smrzka OW, et al. (2000) The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat Genet 25: 19-21.
-
Suslov VV, Ponomarenko PM, Ponomarenko MP, Drachkova IA, Arshinova TV, et al. (2010) TATA box polymorphisms in genes of commercial and laboratory animals and plants associated with selectively valuable traits. Genetika 46: 448-457.
-
Park KK, Jung E, Chon SK, Seo M, Kim HW, et al. (2003) Finding of TRE (TPA responsive element) in the sequence of human taurine transporter promoter. Adv Exp Med Biol 526: 159-166.
-
Seidl CI, Stefan H Stricker, Denise P Barlow (2006) The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. EMBO J 25: 3565-3575.
-
Fatica A, Bozzoni I (2014) Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15: 7-21.
-
Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, et al. (2013) Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9: e1003470.
-
Kelley D, Rinn J (2012) Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol 13: R107.
-
Latos PA, Pauler FM, Koerner MV, Şenergin HB, Hudson QJ, et al. (2012) Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338: 1469-1472.
-
Santoro F, Mayer D, Klement RM, Warczok KE, Stukalov A, et al. (2013) Imprinted Igf2r silencing depends on continuous Airn lncRNA expression and is not restricted to a developmental window. Development 140: 1184-1195.
-
Farmer WT, Farin PW, Piedrahita J A, Bischoff S R, Farin CE (2013) Expression of antisense of insulin-like growth factor-2 receptor RNA non-coding (AIRN) during early gestation in cattle. Anim Reprod Sci 138: 64-73.
-
Farin CE, Farmer WT, Farin PW (2010) Pregnancy recognition and abnormal offspring syndrome in cattle. Reprod Fertil Dev 22: 75-87.





