Journal of Infectious Diseases and Epidemiology
Fluorescent DNA Aptamer-Magnetic Bead Sandwich Assays and Portable Fluorometer for Sensitive and Rapid Foodborne Pathogen Detection and Epidemiology
John G. Bruno*, Taylor Phillips, Alicia M. Richarte, Tiffany Montez, Adrian Garcia and Jeffrey C. Sivils
Pronucleotein Biotechnologies Corporation, San Antonio, TX, USA
*Corresponding author:
John G. Bruno, Pronucleotein Biotechnologies Corporation, 4100 NW Loop 410, Suite 110, San Antonio, TX 78229, USA, Tel: 1-210-731-0015, ext. 2228, Fax: 1-210-731-0041, E-mail: brunobiotech@gmail.com
J Infect Dis Epidemiol, JIDE-2-011, (Volume 2, Issue 1), Research Article; ISSN: 2474-3658
Received: February 06, 2016 | Accepted: March 25, 2016 | Published: March 28, 2016
Citation: Bruno JG, Phillips T, Richarte AM, Montez T, Garcia A, et al. (2016) Fluorescent DNA Aptamer-Magnetic Bead Sandwich Assays and Portable Fluorometer for Sensitive and Rapid Foodborne Pathogen Detection and Epidemiology. J Infect Dis Epidemiol 2:011. 10.23937/2474-3658/1510011
Copyright: © 2016 Bruno JG, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
This report extends fluorescent DNA aptamer-magnetic bead-based sandwich assays developed and published for ultrasensitive detection of Campylobacter jejuni and Listeria monocytogenes by adding prototype assays for pathogenic Escherichia coli, Shiga-like toxin-1 and Salmonella enterica. These tests are assessed by a highly portable fluorometer, thus enabling on-site detection and tracking or epidemiology of major foodborne pathogen outbreaks potentially from "farm to fork". New simplified sandwich assays having fewer steps and employing either aptamer-quantum dot or aptamer-peroxidase conjugate fluorescence reporters are presented along with titration curves to establish approximate limits of detection for E. coli O157 and S. enterica of ≤ 100 cfu/ml and ~100 pg/ml for Shiga-like toxin-1 in buffer. In addition, timed studies in culture enrichment broth suggest detection of E. coli O157 within 4 hours after an inoculum of ~ 10 cfu per 200 ml of broth. Finally, data are presented suggesting consistent detection of E. coli O157 and S. enterica in enrichment cultures for baby spinach, beef trim and diced tomatoes which were processed according to standard FDA BAM methods. Taken together, these data support further development and implementation of a menu of very sensitive, rapid (30 minute) and highly portable aptamer-based fluorescence assays for facile detection of the major foodborne pathogens or their toxins on-site or in central labs to combat the growing health and legal issues associated with foodborne illness and to curb or halt outbreaks during their occurrence.
Keywords
Amplex Red, Aptamer, E. coli, foodborne, Fluorescence, Magnetic bead, Portable, Resazurin, Salmonella, Shiga toxin
Introduction
Food safety continues to be a major public health concern with recent high profile pathogen outbreaks at Blue Bell ice cream and Chipotle restaurants illustrating the huge financial implications for consumers and food producers, distributors, and even restaurant chains. The food safety testing industry already has well-developed confirmatory tests based on post-culture enrichment bacterial and viral genetics, PCR, etc. in central laboratories which are quite necessary, but generally non-portable and too slow to enable rapid detection of pathogens on-site along the food supply chain. In addition, only a very small fraction of domestic and imported foods are sampled and tested, despite the mandate for increased testing inherent in the Food Safety Modernization Act (FSMA) of 2011. Thus, it appears that the food safety testing industry and government inspectors could truly benefit from more rapid and portable, yet highly sensitive and specific screening tests for presumptive detection and tracking or epidemiology of pathogens or their biotoxins. Indeed, examination of recent literature seems to indicate a trend towards more rapid and portable food safety screening technologies [1-4]. Improved screening tests should include testing of food processing equipment and surfaces as well as the foods themselves. Thus, Pro nucleotein, Inc. has been developing ultrasensitive (5-100 bacteria per ml) and rapid (30 minute or less) DNA aptamer-magnetic bead (MB)-based tests for the major foodborne pathogens and biotoxins coupled with the highly portable handheld "FLASH reader" [5-7] to enhance food safety screening capabilities especially in the field.
Interest in development and application of DNA aptamers as surrogates for antibodies in various microbial assays [8,9], including foodborne pathogen detection diagnostics [10-28] has increased dramatically over the last decade. Reasons for the increased research interest in aptamers stem from aptamer's obviating of animal hosts [29] and potentially greater affinity versus antibodies [29] which translates into greater assay sensitivity. In addition, aptamers exhibit greater reproducibility from batch-to-batch versus antibodies [30,31] due to the extremely high fidelity chemical synthesis of DNA [29] leading to more consistent assays.
Our group has already published ultrasensitive detection of Campylobacter jejuni in chicken rinsate using an aptamer-MB plus aptamer-quantum dot conjugate sandwich assay [5] which was confirmed by the U.S. Army Research Laboratory using the same aptamers [25]. We also reported the ultrasensitive detection of Listeria species in culture enrichment broth using an aptamer-MB plus aptamer-peroxidase conjugate sandwich assay and Amplex Ultra RedTM (AUR; resazurin derivative) [6], because it was more reliable than the quantum dot approach in a variety of fatty food matrices. Here we report similar highly sensitive aptamer-MB sandwich assay results for the detection of E. coli O157 and Shiga-like toxin-1 (Stx-1) produced by pathogenic E. coli serotypes such as O157:H7 and other pathogenic E. coli serotypes [32]. We completed our initial assay menu with an aptamer assay for detection of Salmonella enterica serovar Typhimurium in buffer and various food or enrichment broth matrices.
The data reported herein were obtained by using a simplified sandwich assay approach involving fewer steps than similar previously reported assays (Figure 1A). This repertoire of assays coupled with the highly portable FLASH reader (Figure 1B) enable rapid (~ 30 minutes) and effective on-site screening of foodborne threats in and on various foods and fomites. Our system may therefore aid in prevention of illness and possible death from food products and the subsequent economic losses associated with expensive food recalls, lawsuits, or potential bankruptcies for food producers found liable of food safety infractions. As a result, this rapid, ultrasensitive and specific field screening technology led to our company (Pronucleotein, Inc.) winning second place among 49 academic and industrial competitors in the 2014 FDA Food Safety Testing Technology Challenge [33-35] attesting to the potential significance of the technology for future on-site screening, tracking (epidemiology) and reduction or amelioration of foodborne illness outbreaks.
Materials and Methods
Bacteria, shiga-like toxin, DNA and other key materials
E. coli O157:H7 and Salmonella enterica serovar Typhimurium (ATCC 13311) were purchased from American Type Culture Collection (ATCC; Manassas, VA) grown on blood agar plates at 37°C and suspended in sterile phosphate buffered saline (PBS, pH 7.2) for experiments. Shiga-like toxin-1 (Stx-1 from E. coli) was purchased from List Biological Laboratories, Inc. (Campbell, CA). Recombinant intimin protein was purchased from My Bio source, Inc. (San Diego, CA). All aptamer DNA templates, 5'-biotinylated aptamers, and primers were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). Streptavidin (SAv)-coated and tosyl-M270 (2.7 μm) paramagnetic beads (MBs), red streptavidin-coated quantum dots (SAv-Qdot 655), and Amplex Ultra Red™ (AUR) were purchased from Life Technologies, Inc. (Carlsbad, CA). SAv-peroxidase conjugate (~ 1 mg/ml) was purchased from Thermo Fisher, Inc. (Pittsburgh, PA).
DNA aptamer development
Eight to ten rounds of aptamer affinity selection and PCR amplification were performed according to the authors' published procedures [5-7] against both whole bacterial cells and tosyl-MB-immobilized intimin and Stx-1 proteins. After 72 base aptamers were down selected from the random library, they were cloned using a Lucigen GC cloning kit (Middleton, WI) and sent for DNA sequencing at Seque Tech, Inc. (Mountain View, CA). Aptamer DNA sequences were then analyzed for full-length consensus sequences or regions of homology. All candidate aptamer sequences were synthesized with a 5'-biotin moiety and then used in ELISA-like (ELASA) assays to screen for the highest affinity and most specific aptamer assay candidates according to published protocols [5-7]. The actual aptamer DNA sequences cannot be divulged here because of their potential commercial value. However, the aptamer DNA sequences employed in the reported assays are a subset of the company's U.S. patent-pending aptamer DNA sequence library.
Simplified Aptamer-MB sandwich assay procedure
To reduce the number of steps in a typical aptamer-MB sandwich assay (Figure 1A), the authors began combining 500 pmoles of 5'-biotinylated aptamers and the target bacteria in 500 μl of 1X B&W (binding and washing) buffer (1M NaCl, 5 mM Tris-HCl and 0.5 mM EDTA) followed by gentle mixing for 10 min at room temperature. Then 25 μl of 1:10 diluted stock M270-SAv-MBs in 1X B&W were added to the tube containing the 5-biotinylated aptamers and bacteria and the tube was again gently mixed for 10min at room temperature. The tube was then placed on a Dynal magnetic collection rack for ~ 1 minute collection of the MBs as a small pellet. Thereafter, the MB pellet was washed by gently re suspending in 1 ml of 1X B&W + 0.01% Tween 20. After washing and resuspension in 500 μl of 1:100 diluted SAv- peroxidase conjugate (~ 10 ng) Amplex Ultra Red™ (AUR) was prepared and activated with H2O2 and fluorescence readings were taken with the FLASH reader as previously described [6,7]. In some cases, aptamer-5'-biotin-SAv-Qdot 655 conjugates were prepared, purified through Sephadex G25 columns and used in place of the aptamer- peroxidase reporter reagent and AUR (Figure 1A) followed by assessment in the FLASH reader and by fluorescence microscopy as previously described [5-7]. Stx-1 sandwich assays were similarly conducted using aptamer-Qdot 655 conjugate reporters tittered against various levels of purified Stx-1 protein spiked into PBS.
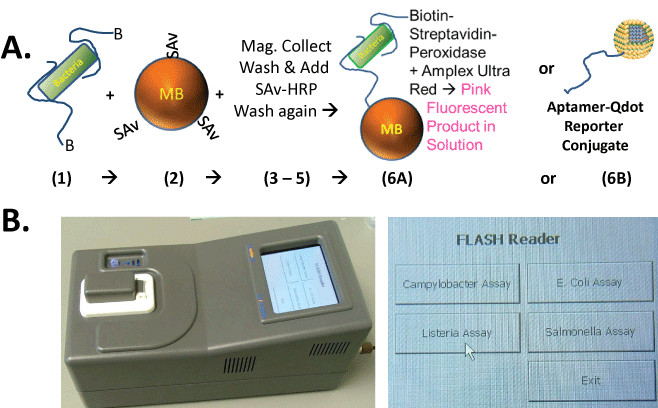
.
Figure 1: (A) Diagram of the simplified sandwich assay protocol in which biotinylated capture and reporter aptamers which may or may not be the same are added to the sample containing pathogenic bacteria and allowed to bind (step 1), followed by addition of streptavidin (SAv)-coated magnetic microbeads (MBs) in step 2, magnetic collection, washing, and addition of SAv-peroxidase or SAv-quantum dots (Qdots) and washing in steps 3-5, followed finally by fluorescence development with Amplex Ultra Red™ (AUR; resazurin derivative) and peroxide or detection of Qdot fluorescence (steps 6A or 6B); (B) Appearance of the handheld, rechargeable, FLASH reader fluorometer with an on board computer and touch screen menu of various key foodborne pathogen assays.
View Figure 1
Timed enrichment culture studies
For the timed enrichment culture studies, an average of 10 E. coli O157:H7 cells were inoculated (spiked) into 200 ml of sterile Tryptic Soy Broth (TSB) in sterile 400 ml Erlenmeyer flasks plugged with sterile cotton and cultured at 37°C with gentle shaking for up to 4 hours. 3 × 1 ml samples were drawn from two separate spiked and control (negative blank) shaker flasks each hour for 4 hours for the simplified sandwich assay and fluorescence assessment using the FLASH reader. In addition, 100 μl samples were drawn hourly from the flasks for spread plate counts on blood agar plates cultured at 37°C overnight to verify that the E. coli O157:H7 culture was growing and to determine approximately how many cells were detected each hour.
FDA-compliant BAM methods used to enrich for spiked pathogens in various food samples
Using the FDA standard Bacteriological Analytical Manual (BAM) protocols, enrichment of baby spinach leaves, beef trim and canned diced tomato esspiked with E. coli O157:H7 or Salmonella enterica ATCC 13311, several of the best DNA aptamer sandwich assay combinations were conducted. The FDA-BAM method for Salmonella detection was used for diced canned tomatoes which were enriched in Buffered Peptone Water as per the BAM method instructions. After both pre-enrichment steps for Salmonella, the pH was checked and adjusted to the required pH as described by the BAM for Salmonella. Baby spinach and beef trim were processed in Lactose Broth, again as per the BAM directions. Tetrathionate (TET) Broth and Rappaport-Vasilliadis (RV) Broth along with Iodine and Brilliant Green dye supplements for the TET were used as well. One ml samples were transferred to TET and 100 μL to RV as per the directions. The FDA-BAM method for E. coli was closely followed as well. All matrices were pre-enriched in Brain Heart Infusion Broth for 3 hours as per the BAM directions and then diluted with 2X Tryptone Phosphate Broth to make a 1X enrichment medium as per the BAM protocol. This material was incubated for 20 hours and then streaked to Eosin Methylene Blue and Mac Conkey's agar plates. The concentrations of bacteria detected in the various food matrices were later confirmed to be ~ 380,000 E. coli O157:H7 per ml and ~ 1.04 million S. enterica 13311 ml by spread plate counts.
Results
Figure 1A illustrates the simplified sandwich assay format and protocol. It is noteworthy that one can add the 5'-terminal bio tinylated aptamers to a sample containing bacteria and allow the capture aptamers to bind bacterial surfaces followed by addition of SAv-coated MBs instead of first coating the SAv-MBs with bio tinylated aptamers and then adding the aptamer-coated MBs to the bacterial sample. This action saves at least one wash step in which the aptamer-coated MBs would need to be collected with a magnet and then washed free of any excess capture aptamers not bound to the MBs before proceeding to probing of the food sample for pathogenic bacteria. Another step is saved because the bio tinylated aptamers bound to the bacterial cells, but not to the MB surface, are free to act as reporter aptamers when SAv-peroxidase or SAv-Qdot conjugates are added. This again simplifies the assay by obviating the addition of a separate reporter aptamer with associated wash steps. Figure 1B illustrates the outward appearance of the handheld FLASH reader and its touch screen menu of various foodborne pathogen assays.
Figure 2 illustrates that unlike whole bacterial cell capture for their previous published Campylobacter and Listeria aptamer-MB sandwich assays [5,6], under the fluorescence microscope the authors observed primarily speckled and punctate coating on the aptamer-MBs exposed to E. coli O157:H7 and S. enterica. These punctate staining patterns, suggest that the aptamer-MBs are actually capturing released or secreted E. coli intimin proteins and S. enteric outer membrane proteins (OMPs) [36] or outer membrane vesicles [37] instead of whole cells during magnetic collection and washing. Regardless of which parts of these pathogenic bacteria (whole cells, OMPs or outer membrane vesicles) are captured, the photomicrographs still strongly suggest positive identification of the pathogens presence based upon detection of the targeted proteins (Figure 2B and Figure 2D), because the negative control blanks without added bacteria (Figure 2A and Figure 2C) are essentially dark and non-fluorescent with only a slight nonspecific background fluorescence which may be due to low level aptamer-biotin binding to unoccupied streptavidin sites on the surface of the Qdots in the absence of bacteria.
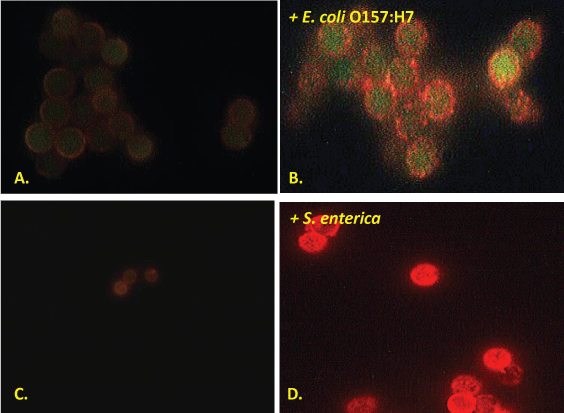
.
Figure 2: Typical fluorescence photomicrographs of negative control fluorescence for the E. coli intimin and S. enterica aptamer-MB plus aptamer-Qdot 655 assays (panels A and C respectively) and the same assays conducted in the presence of ~ 106 live E. coli O157:H7 or S. enterica 13311 bacteria per ml (panels B and D respectively) showing punctate speckled red fluorescence suggestive of aptamer-magnetic bead capture of released or secreted outer membrane proteins [36] or vesicles [37] from live Gram negative bacterial cells.
View Figure 2
Data in Figure 3A suggest a probable detection limit of less than 100 E. coli O157:H7 per ml in buffer using the intimin sandwich assay. Figure 3B suggests that a different general E. coli assay can detect E. coli O157:H7 as early as 4 hours post-inoculation of an average of 10 cfu per 200 ml of TSB in a shaker culture, thus significantly cutting detection time in central food testing laboratories.
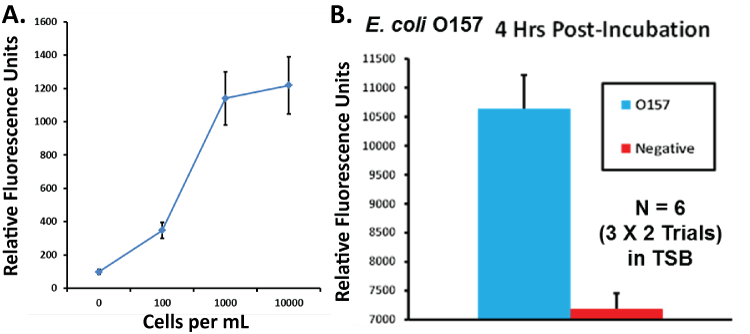
.
Figure 3: (A) Intimin aptamer-MB plus aptamer-peroxidase AUR sandwich assay titration curve versus several logs of E. coli O157:H7 per ml in PBS showing a probable detection limit of ≤ 100 bacterial cells per ml. Data points represent the means and standard deviations of three independent trials; (B) Results of a timed enrichment study utilizing a general E. coli aptamer-MB plus aptamer-peroxidase AUR assay for E. coli O157:H7 demonstrating detection from a tryptic soy broth enrichment culture at 4 hours post-inoculation of ~ 10 viable E. coli O157:H7 into 200 ml of TSB in a shaker flask culture.
View Figure 3
Figure 4 illustrates that although the Stx-1 aptamer-MB plus aptamer-Qdot 655 sandwich assay hooks somewhat at higher concentrations of Shiga-like toxin 1 in buffer, which is common especially in simplified or one-step sandwich assays [38,39], it is still capable of detecting 0.1 ng or 100 pg/ml or less of the toxin. The high dose hook effect is not problematic because the fluorescence intensity values at 1-100 ng/ml in Figure 4 are still well above the blank levels (i.e., would register as positive detection events). In addition, even if problematic (leading to false negative results), such hook effect could always be overcome by diluting samples and running the assay in the linear dynamic range [40]. Stx-1 detection would provide a valuable adjunct assay to the intimin assay to detect most of the pathogenic E. coli serotypes [32,41].
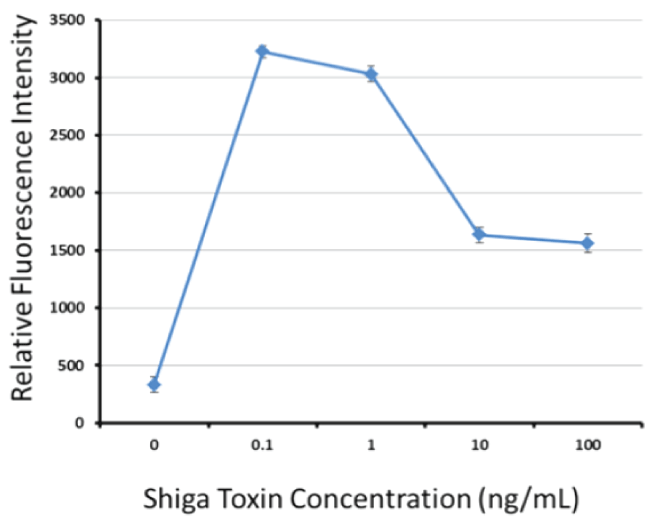
.
Figure 4: Titration results of the best Stx-1 aptamer-MB plus aptamer-Qdot 655 sandwich combination versus varied levels of Stx-1 in PBS showing a detection limit of ≤ 0.1 ng or 100 pg/ml and a nonlinear dynamic range of ≤ 0.1 to 100 ng/ml despite some "hook" effect at the higher Stx-1 concentrations. Data points represent the means and standard deviations of three independent trials.
View Figure 4
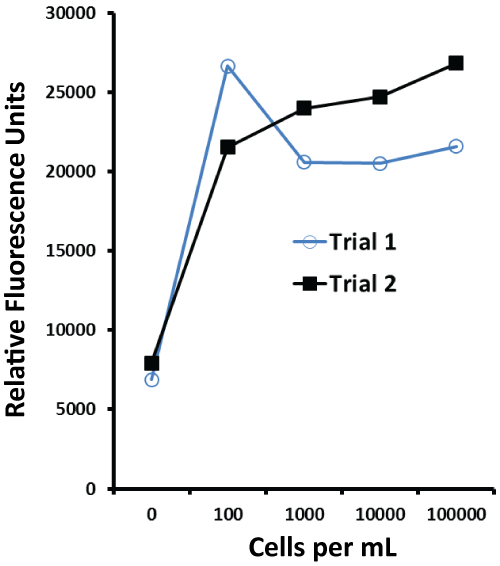
The independent titration curves shown in Figure 5 illustrate that the aptamer-MB plus aptamer-peroxidase AUR assay was able to detect less than100 S. enteric 13311 cells per ml in buffer and had a dynamic range of at least 100 to 100,000 cells per ml.
.
Figure 5: Results of two independent aptamer-MB plus aptamer-peroxidase AUR trials for the Salmonella whole cell outer membrane protein (OMP) assay showing a detection limit of < 100 cells per ml and dynamic range of < 100 to 100,000 S. enterica 13311 cells per ml.
View Figure 5
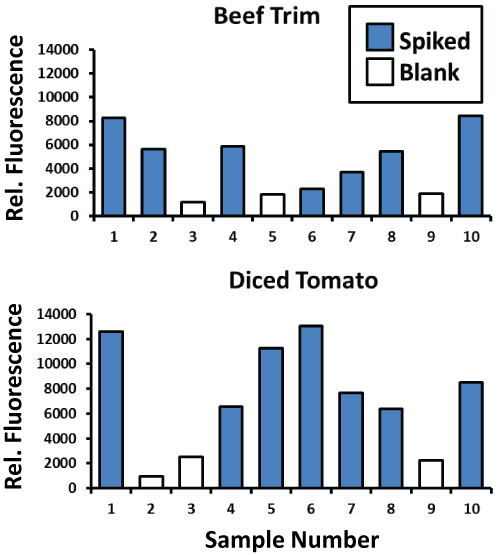
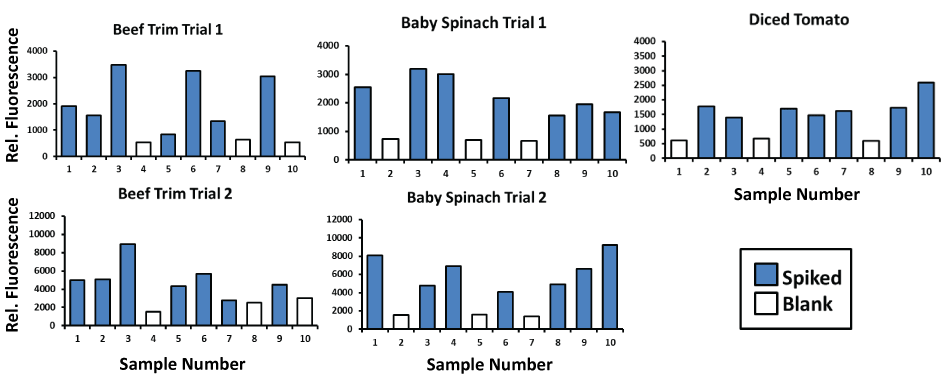
When applied to enriched beef trim and diced tomato samples (Figure 6) processed according to the FDA's BAM methods, the authors' general E. coli aptamer-MB plus aptamer- peroxidase AUR assay consistently detected ~ 380,000 E. coli O157:H7 cells per ml (blue bars) in these enrichment matrices versus the consistently low background fluorescence of the unspiked blank samples (white bars). A second set of trials for the S. enterica aptamer-MB plus aptamer- peroxidase AUR assay consistently detected ~ 1.04 million S. enterica cells per ml in these enriched samples (Figure 7) of beef trim, baby spinach and diced tomatoes. The data in Figure 6 and Figure 7 are key to illustrating that the aptamer-based assays can function in real enriched food samples for central lab testing as well as field-portable screening assays.
.
Figure 6: Results for general E. coli OMP aptamer-MB plus aptamer-peroxidase AUR detection of E. coli O157:H7 bacteria at ~ 380,000 per ml in beef trim and diced tomato enriched samples performed according to the standard FDA BAM method (see Materials and Methods). Unspiked blank samples are represented by the white bars and test samples spiked with E. coli O157:H7 are represented by blue bars.
View Figure 6
.
Figure 7: Results for general S. enterica OMP aptamer-MB plus aptamer-peroxidase AUR detection of S. enterica 13311 bacteria at ~ 1.04 million per ml in beef trim, baby spinach, and diced tomato enriched samples performed according to the standard FDA BAM method (see Materials and Methods). Unspiked blank samples are represented by the white bars and test samples spiked with S. enterica 13311 are represented by blue bars.
View Figure 7
Discussion
Detection of minute levels of foodborne pathogens or their secreted toxins in hundreds of milliliters of enrichment broth in well-equipped central testing laboratories is still a difficult, slow (days long) and uncertain process prone to false negative results. Yet, the food safety testing community's system of food safety and foodborne pathogen epidemiology relies on central laboratory testing which only tests a very small fraction of domestic and imported foods. In order to improve foodborne pathogen detection and enhance overall food safety, the industrial and government communities need better (faster, more sensitive and yet accurate or specific) detection tools (sensors and assays) for portable on-site testing of foods and fomites all along the food supply chain literally from "farm to fork".
The authors' simplified aptamer-MB-based fluorescent assays whether based on Qdots or AUR as reported herein and previously [5-7] are an attempt to fuse the high affinity, specificity and reproducibility of aptamers [29] versus traditional antibodies [30,31] with the concentrating and purifying power of MBs and the high sensitivity of fluorescence methods. Although, not indicated in the text, the authors have ample evidence of negative control aptamers in the form of many failed assay attempts using other aptamer DNA sequences during the development phases. Also not shown in the text is the fact that the authors are adapting the aptamer-MB fluorescence assays to sterile wetted sponge swab (SpongesicleTM) samples taken off of inanimate surfaces to rapidly test food grinding and other processing equipment, conveyor belts, etc. on-site. The addition of a very sensitive handheld fluorometer such as the FLASH reader (Figure 1B) provides portability to read the rapid (~ 30 minute) assays described herein and completes the technology package for rapid on-site tracking and epidemiology of foodborne pathogen outbreaks. Although, more research and advanced development is clearly needed, the data presented in this report suggest that aptamer-MB-based fluorescence assays may someday hopefully ameliorate or halt foodborne illness outbreaks. Finally, the same aptamer-MB fluorescence assay technology can be extended beyond food safety applications into the realm of detecting other infectious diseases including parasites [7] or arboviruses [42] and other clinical diagnostic biomarkers or analytes [43-45] to aid in diagnosing and tracking human diseases.
Acknowledgments
Funding was provided by the Texas Emerging Technology Fund, internal funding from Pro nucleotein Biotechnologies, Inc. and second place prize money ($120,000) from the FDA's 2014 Food Safety Testing Technology Challenge.
References
-
Ayvaz H, Rodriguez-Saona LE (2015) Application of handheld and portable spectrometers for screening acrylamide content in commercial potato chips. Food Chem174:154-162.
-
Bülbül G, Hayat A, Andreescu S (2015) Portable nano particle-based sensors for food safety assessment. Sensors 15: 30736-30758.
-
Srinivasan B, Tung S (2015) Development and applications of portable biosensors. J Lab Autom 20: 365-389
-
Zhang Z, Tang X, Wang D, Zhang Q, Li P, et al. (2015) Rapid on-site sensing aflatoxin B1 in food and feed via a chromatographic time-resolved fluoroimmunoassay. PLoS One10: e0123266.
-
Bruno JG, Phillips T, Carrillo MP, Crowell R (2009) Plastic-adherent DNA aptamer-magnetic bead and quantum dot sandwich assay for Campylobacter detection. J Fluoresc 19: 427-435
-
Bruno JG, Phillips T, Montez T, Garcia A, Sivils JC, et al. (2015) Metrix360 Laboratories Development of a fluorescent enzyme-linked DNA aptamer-magnetic bead sandwich assay and portable fluorometer for sensitive and rapid Listeria detection. J Fluoresc 25:173-183
-
Bruno JG, Richarte AM, Phillips T, Savage AA, Sivils JC, et al. (2014) Development of a fluorescent enzyme-linked DNA aptamer-magnetic bead sandwich assay and portable fluorometer for sensitive and rapid Leishmania detection in sandflies. J Fluoresc 24: 267-277
-
Davydova A, Vorobjeva M, Pyshnyi D, Altman S, Vlassov V, et al.(2015) Aptamers against pathogenic microorganisms. Crit Rev Microbiol.10:1-19
-
Torres-Chavolla E, Alocilja EC (2009) Aptasensors for detection of microbial and viral pathogens. Biosens Bioelectron 24:3175-3182.
-
Amaya-González S, de-los-Santos-Álvarez S, Miranda-Ordieres AJ, Lobo-Castañón MJ (2013) Aptamer-based analysis: apromising alternative for food safety control. Sensors 13: 16292-16311
-
Karkkainen RM, Drasbek MR, McDowall I, Smith CJ, Young NWG, et al. (2011) Aptamers for safety and quality assurance in the food industry: detection of pathogens. Int J Food Sci Technol 46: 445-454.
-
Liu X, Zhang X (2015) Aptamer-based technology for food analysis. Appl Biochem Biotechnol 75:603-624
-
Duan N, Wu S, Zhu C, Ma X, Wang Z, et al. (2012) Dual-color upconversion fluorescence and aptamer-functionalized magnetic nanoparticles-based bioassay for the simultaneous detection of Salmonella typhimurium and Staphylococcus aureus. Anal Chim Acta 723: 1-6.
-
Hyeon JY, Chon JW, Choi IS, Park C, Kim DE, et al.(2012) Development of RNA aptamers for detection of Salmonella enteritidis. J Microbiol Methods 89: 79-82.
-
Jyoti A, Vajpayee P, Singh G, Patel CB, Gupta KC, et al.(2011) Identification of environmental reservoirs of nontyphoidal salmonellosis: aptamer-assisted bioconcentration and subsequent detection of Salmonella typhimurium by quantitative polymerase chain reaction. Environ Sci Technol 45: 8996-9002.
-
Khang J, Kim D, Chung KW, Lee JH (2016) Chemiluminescent aptasensor capable of rapidly quantifying Escherichia coli O157:H7. Talanta 147:177-183.
-
Lee SH, Ahn JY, Lee KA, Um HJ, Sekhon SS, et al. (2015) Analytical bioconjugates, aptamers, enable specific quantitative detection of Listeria monocytogenes. Biosens Bioelectron 68: 272-280.
-
Lee YJ, Han SR, Maeng JS, Cho YJ, Lee SW (2012) In vitro selection of Escherichia coli O157:H7-specific RNA aptamer. Biochem Biophys Res Commun 417: 414-420.
-
Ohk SH, Koo OK, Sen T, Yamamoto CM, Bhunia AK (2010) Antibody-aptamer functionalized fibre-optic biosensor for specific detection of Listeria monocytogenes from food. J Appl Microbiol 109: 808-817.
-
Pan Q, Zhang XL, Wu HY, He PW, Wang F, et al .(2005) Aptamers that preferentially bind type IVB pili and inhibit human monocytic-cell invasion by Salmonella enterica serovar typhi. Antimicrob Agents Chemother 49: 4052-4060.
-
Park HC, Baig IA, Lee SC, Moon JY, Yoon MY (2014) Development of ssDNA aptamers for the sensitive detection of Salmonella typhimurium and Salmonella enteritidis. Appl Biochem Biotechnol 174: 793-802.
-
Queirós RB, Gouveia C, Fernandes JR, Jorge PA (2014) Evanescent wave DNA-aptamer biosensor based on long period gratings for the specific recognition of E. coli outer membrane proteins. Biosensors & Bioelectronics. 62: 227-233
-
Moon J, Kim G, Lee S, Park S (2013) Identification of Salmonella typhimurium-specific DNA aptamers developed using whole-cell SELEX and FACS analysis. J Microbiol Methods 95:162-166.
-
Singh G, Vajpayee P, Rani N, Jyoti A, Gupta KC, et al. (2012) Bio-capture of S. typhimurium from surface water by aptamer for culture-free quantification. Ecotoxicol Environ Safety 78: 320-326.
-
Stratis-Cullum DN, McMasters S, Pellegrino PM (2009) Evaluation of relative aptamer binding to Campylobacter jejuni bacteria using affinity probe capillary electrophoresis. Analyt Lett 42:2389-2402.
-
Suh SH, Dwivedi HP, Choi SJ, Jaykus LA (2014) Selection and characterization of DNA aptamers specific for Listeria species. Anal Biochem 459: 39-45.
-
Suh SH, Jaykus LA (2013) Nucleic acid aptamers for capture and detection of Listeria spp. J Biotechnol 167: 454-461.
-
Wu WH, Li M, Wang Y, Ouyang HX, Wang L, et al.(2012) Aptasensors for rapid detection of Escherichia coli O157:H7 and Salmonella typhimurium. Nanoscale Res Lett 7: 658.
-
Jayasena SD (1999) Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem 45: 1628-1650.
-
Bordeaux J, Welsh A, Agarwal S, Killiam E, Baquero M, et al. (2010) Antibody validation. BioTechniques 48: 197-209.
-
Marx V (2013) Finding the right antibody for the job. Nature Methods 10: 703-707.
-
Kaper JB, Nataro JP, Mobely HLT (2004) Pathogenic Escherichia coli. Nature Rev Microbiology 2: 123-140.
-
Anonymous (2015) FDA names first-ever food safety challenge finalists. Food Safety News.
-
Holliman K (2015) Food safety innovations win awards from FDA. Food Quality & Safety.
-
Wu J (2015) FDA's food safety challenge seeks speedier Salmonella detection. US News World Report.
-
Hellman J, Loiselle PM, Tehan MM, Allaire JE, Boyle LA, et al.(2000) Outer membrane protein A, peptidoglycan-associated lipoprotein, and murein lipoprotein are released by Escherichia coli bacteria into serum. Infect Immun 68: 2566-2572.
-
Schwechheimer C, Kuehn MJ (2015) Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13: 605-619.
-
Fernando SA, Wilson GS (1992) Studies of the 'hook' effect in the one-step sandwich immunoassay. J Immunol Methods 151: 47-66.
-
Tate J, Ward G (2004) Interferences in immunoassay. Clin Biochem Rev 25: 105-120.
-
Butch AW (2000) Dilution protocols for detection of hook effects/prozone phenomenon. Clin Chem 46: 1719-1721.
-
Adu-Bobie J, Trabulsi LR, Carneiro-Sampaio MM, Dougan G, Frankel G (1998) Identification of immunodominant regions within the C-terminal cell binding domain of intimin alpha and intimin beta from enteropathogenic Escherichia coli. Infect Immun 66: 5643-5649.
-
Bruno JG, Carrillo MP, Richarte AM, Phillips T, Andrews C, et al.(2012) Development, screening, and analysis of a small DNA aptamer library potentially useful for diagnosis and passive immunity of arboviruses. BMC Res Notes 5: 633.
-
Bruno JG, Richarte AM (2016) Development and characterization of an enzyme-linked DNA aptamer-magnetic bead-based assay for human IGF-I in serum. Microchem J 124: 90-95
-
Bruno JG, Richarte AM, Phillips T (2014) Preliminary development of a DNA aptamer-magnetic bead capture electrochemiluminescence sandwich assay for Brain Natriuretic Peptide. MicrochemJ 115: 32-38.
-
Vance SA, Sandros MG (2014) Zeptomole detection of C-Reactive Protein in serum by a nanoparticle amplified surface plasmon resonance imaging aptasensor. Sci Reports 4: 5129.





