International Journal of Cancer and Clinical Research
Common Polymorphisms in the USF1 Gene and Cancer Susceptibility: A Meta-Analysis
Meng Zhang1#, Jian Bai11#, Junjie Huang2#, Yukun Ge6, Hu Xiong5, Wei Lu1, Jizhou Shi8, Lu Fang5, Song Wu1,3,7* and Zhiming Cai1
1Department of Urology, Shenzhen Second People's Hospital, clinical medicine college of Anhui Medical University, Shenzhen, China
2School of Medicine, Shenzhen University, Shenzhen, China
3Department of Tumor Immunology, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou, China
4Department of Urology, The second Hospital of Lanzhou University, Lanzhou, China
5Department of Urology, The second affiliated hospital of Anhui Medical University, Hefei, China
6Department of Urology, Zhujiang Hospital, Southern Medical University, Guangzhou, China
7Luohu District People's Hospital, Shenzhen, China
8Department of Medicine, Shengli Oilfield Central Hospital, Dongying, Shandong, China
#Authors contributed equally
*Corresponding author:
Song Wu, Shenzhen Second People's Hospital, clinical medicine college of Anhui Medical University, Shenzhen Guangdong, China, E-mail: Songwu2004@yeah.net
Int J Cancer Clin Res, IJCCR-2-025, (Volume 2, Issue 4), Original Article; ISSN: 2378-3419
Received: July 19, 2015 | Accepted: September 19, 2015 | Published: September 21, 2015
Citation: Zhang M, Bai J, Huang J, Ge Y, Xiong H, et al. (2015) Common Polymorphisms in the USF1 Gene and Cancer Susceptibility: A Meta-Analysis. Int J Cancer Clin Res 2:025. 10.23937/2378-3419/2/4/1025
Copyright: © 2015 Zhang M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Upstream transcription factor 1(USF1) has been identified to be implicated in the development of many cancer categories. In view of recent studies, several polymorphisms in USF1 gene appeared to exert diverse influence on cancer susceptibility. However, the association between USF1 polymorphisms and cancer susceptibility remains inconclusive due to the finite relevant published discoveries. Therefore, we conducted a meta-analysis by pooling all available published data on the susceptibility of USF1 (rs2516838, rs2516839, rs2774276 and rs3737787) polymorphisms to cancer. The pooled odds ratios (ORs) with 95% confidence intervals (95% CIs) were calculated. Finally, nine independent case-control studies comprising of 1,927 cases and 4,037 controls were enrolled in the present study. An increased susceptibility was identified inrs2516839 polymorphism and rs3737787 polymorphism, whereas no association was identified in rs2516838 andrs2774276.In addition, a statistically significant association between the USF12516839 polymorphism and HCC was revealed. Nonetheless, more individual studies with higher quality are required for further elucidation.
Keywords
USF1, Polymorphism, Cancer, Meta-analysis
Introduction
The etiology of cancer is obscure because of the involvement of multiple risk factors including complicated gene-gene and gene-environment interactions. Hepatocellular carcinoma (HCC) is one of the most occurring cancers and a leading cause of cancer-related deaths worldwide, especially in China [1]. Currently, the well-recognized risk factors for HCC include chronic viral hepatitis (HBV, HCV), smoking, alcohol consumption, aflatoxin exposure and liver cirrhosis [2,3]. Among these reasons, chronic inflammation plays a crucial role in the development of cancer [4,5]. Increasing evidence suggests that single nucleotide polymorphisms (SNPs) in genes are responsible for key elements of chronic inflammation development and progression, which may lead to HCC as well as papillary thyroid cancer (PTC) [6,7]. SNP is one of the most common types of genetic abnormalities. Recently, polymorphisms have become a hot spot of research, which is expected to be the molecular markers for predicting disease susceptibility and ponderable in individual treatment. Increasing attention has been paid to genetic susceptibility and several candidate genes have been identified in recent years.
Upstream transcription factor 1 (USF1), which is located on chromosome 1q22-23, is an important transcription factor in human genome. It belongs to the helix-loop-helix leucine zipper family. USF1 functions as a ubiquitously expressed transcription factor that regulates gene transcription by binding to the E-box motif of target genes [8]. It is engaged in the transcription activation of various functional genes implicated in different physiological processes, such as lipid and glucose metabolism [9,10], stress response [11], immune response [12,13], cell cycle control [14] and tumor suppression [15]. Accordingly, genetic variations of USF1 may be correlated with some metabolic syndromes and cardiovascular diseases such as familial combined hyperlipidemia (FCHL) [16,17] high plasma triglyceride [18], low-density lipoprotein (LDL) level [19], atherosclerosis lesions [20], coronary artery calcifications [21], and low APOE expression [16].
As for the risk of cancer, some case-control studies in USF1 were performed among Chinese population. According to the previous studies, two polymorphisms in USF1 (rs2516839 and rs3737787) have been demonstrated to be associated with HCC susceptibility in Chinese [22,23]. While in another research three polymorphisms in USF1 (rs2516838, rs3737787 and rs2516839) showed significant association with PTC susceptibility in Chinese population [24]. Nonetheless, the association of USF1 polymorphisms and cancer susceptibility remains an issue due to inconclusive findings in currently published case-control studies. Therefore, the present study was carried out to investigate the effect of USF1 polymorphisms on cancer susceptibility by pooling all currently available data.
Materials and methods
Search strategy
A comprehensive literature search was performed in PubMed, Embase, Web of Science databases (up to 15 June 2015) to collect all eligible studies on the relevance between polymorphisms of USF1 (rs2516838, rs2516839, rs2774276 and rs3737787) and cancer susceptibility by using the following search strategy: ("USF1"or "upstream transcription factor 1") and ("polymorphism "or" mutation") and ("cancer "or" tumor"). Moreover, studies were identified by a manual search of the reference lists of eligible reviews and retrieved studies for additional studies.
Inclusion criteria
The articles adopted in our current meta-analysis met the following criteria: (a) studies that evaluated the relevance between the polymorphisms in USF1 and cancer susceptibility; (b) case-control study; (c) an OR with a 95% can be obtained from all the cases and controls. We excluded studies which were: (a) case-only studies and Reviews; (b) insufficient raw statistics to calculate odds ratios (ORs) with 95% confidence intervals (CIs); (c) duplicated publications; (d) studies based on families.
Data extraction
Three investigators (M.Zhang, J.Bai and J.J. Huang) extracted the data independently from adopted studies and a consensus was reached on every item. Any disagreement was settled according to the description above. The following statistics was collected: first author's surname, publication year, ethnic population, sample size of cases and control, source of controls, and genotype or allele distribution in cases and controls.
Statistical analysis
ORs and 95% CIs in the case-control studies were employed to assess the association between the USF1 polymorphisms and cancer susceptibility. The pooled ORs were performed under the allele contrast(C vs. T), dominant (CC+TC vs. TT), and recessive (CC vs. TC+TT) models. Comparisons were made in heterozygote (TC vs. TT) and homozygote (CC vs. TT). The P values of HWE for the genotype distribution in controls were calculated by χ2 test. The meta-analyses were conducted using the STATA 12.0 (Stata Corporation, College Station, Texas). A chi-square based Q-statistic test was conducted for the heterogeneity of studies within the case-control studies [25]. If the Q test (P > 0.1) presented homogeneity in studies, we would applied the fixed effects model [26]; Otherwise, the random effects model was selected [27]. The inconsistency index was also adopted to evaluate the heterogeneity across studies (I2 > 50%: significant heterogeneity; I2=25-50%: moderate heterogeneity; I2 < 25%: no heterogeneity). Stratification analyses were performed by cancer type and source of control. Sensitivity analysis was conducted by extracting a study each time to evaluate the stability of the results. Begg's funnel plot and Egger's regression test were applied to assess the possibility of publication bias.
Results
The identification and characteristics of eligible studies
After a systematic literature search in the databases of PubMed, Embase and Web of Science on the association between USF1 polymorphisms and cancer including nine independent case-control studies. A total of 1,927 cases and 4,037controls were enrolled in our meta-analysis [22-24]. We presented a flow chart of the studies screening process in Figure 1.They were published between 2014 and 2015.Table1 shows the characteristics of all eligible studies [22-24]. Among the eligible nine case-control studies, three are TaqMan assay, while six are performed by PCR. In addition, two of these case-control studies are population-based and seven are hospital-based. The ethnicity in these case-control studies is Chinese.
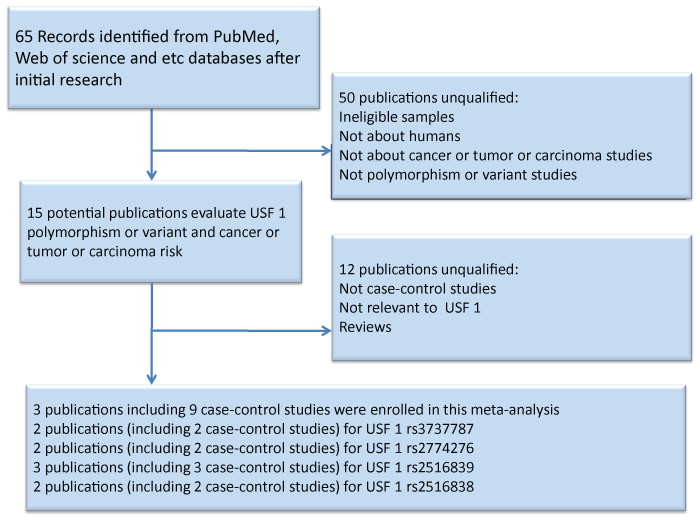
.
Figure 1: Flow chart showing the study selection process. Finally, 3 publications were retrieved reporting a total of 1,927 cases and 4,037 controls.
View Figure 1
![]()
Table 1: Polymorphisms and characteristics of studies involved in this meta-analysis
View Table 1
Association between USF1 polymorphisms and cancer susceptibility
We summarized the main results of the present meta-analysis and the heterogeneity test in Table 2. By pooling ORs and 95 % CIs, it demonstrated that neither USF1 rs2516838 nor USF1 rs2516838 polymorphisms was associated with the susceptibility of cancer.
![]()
Table 2: Results of meta-analysis for polymorphisms in USF1 and cancer susceptibility
View Table 2
Nevertheless, we identified an increased susceptibility in the polymorphism of USF1rs2516839 (C vs. T: OR=1.392, 95%CI=1.203-1.612, Pheterogeneity = 0.113; CC vs. TT: OR = 1.837, 95%CI = 1.386-2.433, Pheterogeneity = 0.243; TC vs. TT: OR = 1.301, 95% CI = 1.017-1.665, Pheterogeneity = 0.904; CC/TC vs. TT: OR = 1.486, 95% CI = 1.185-1.863, Pheterogeneity = 0.594; CC vs. TC/TT: OR = 1.548, 95% CI = 1.219-1.965, Pheterogeneity = 0.155), as well as the polymorphism of USF1 rs3737787 (C vs. T: OR = s1.210, 95%CI = 1.082-1.354, p = 0.421; CC vs. TT : OR = 1.325, 95% CI = 1.003-1.751, Pheterogeneity = 0.622; TC vs. TT: OR = 1.277 95% CI = 1.124-1.450, Pheterogeneity = 0.380; CC/TC vs. TT: OR = 1.227, 95% CI = 1.105-1.362, Pheterogeneity = 0.362). When we stratified the analysis by types of cancer, the polymorphism of USF1 rs2516839 was revealed to contribute to HCC cancer susceptibility and the pooled results were statistically significant (C vs. T: OR = 1.655, 95% CI = 1.284-2.135, Pheterogeneity = 0.192; CC vs. TT: OR = 2.496, 95% CI = 1.529-4.074, Pheterogeneity = 0.445; CC/TC vs. TT: OR = 1.727, 95% CI = 1.184-2.519, Pheterogeneity = 0.749; CC vs. TC/TT: OR = 2.037, 95% CI = 1.324-3.134, Pheterogeneity = 0.04). Additionally, a significant association of the polymorphism of USF1 rs2516839 was found in hospital-based subgroup. (C vs. T: OR = 1.373, 95% CI = 1.164-1.1620, Pheterogeneity = 0.192; CC vs. TT: OR = 1.771, 95% CI = 1.293-2.424, Pheterogeneity = 0.110; CC vs. TC/TT: OR = 1.515, 95% CI = 1.160-1.979, Pheterogeneity = 0.058).
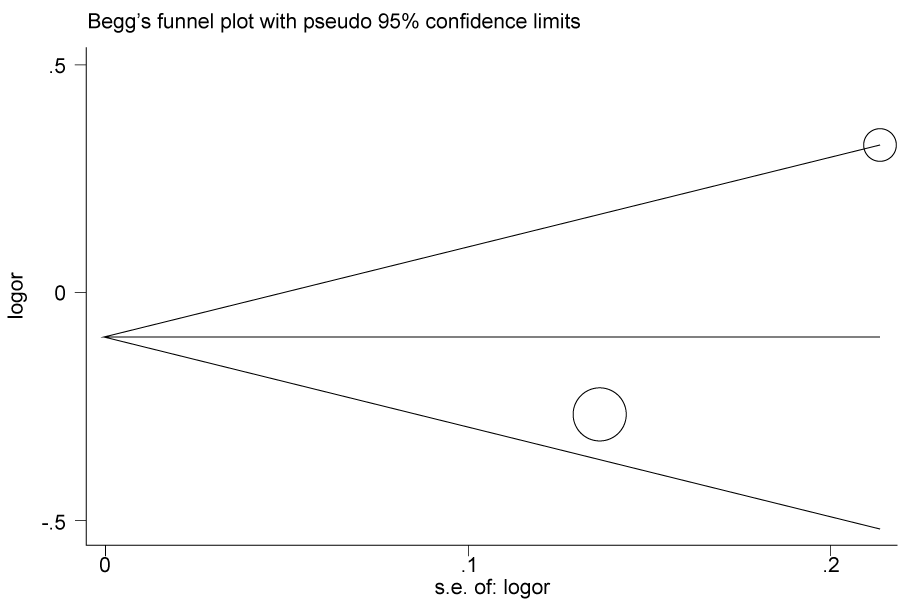
.
Figure 2a: Publication bias in studies of the association between the USF1 rs2516838 polymorphism and cancer susceptibility assessed by Begg's funnel plot and Egger's test (C vs T). Log (OR): the natural logarithm of the odds ratio.
View Figure 2a
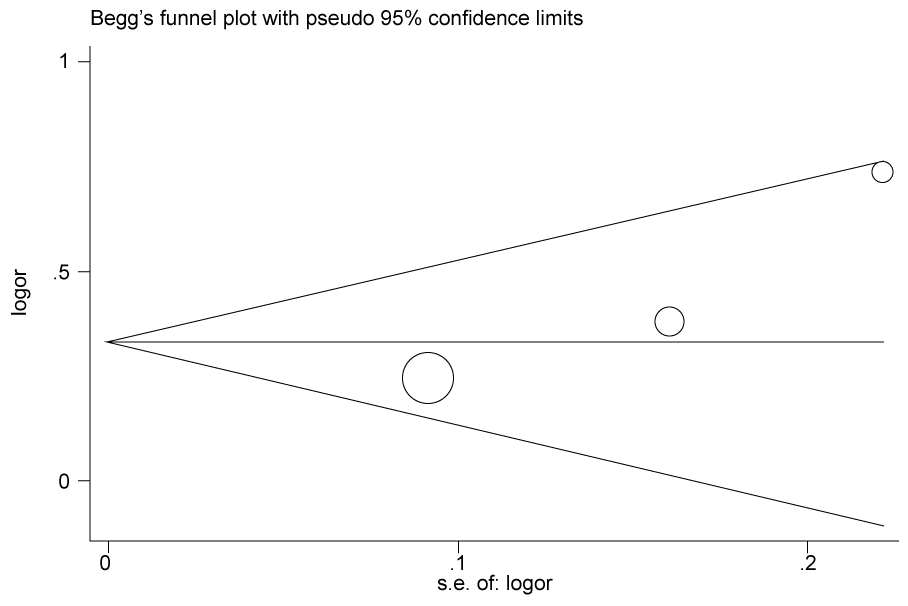
.
Figure 2b: Publication bias in studies of the association between the USF1 rs2516839polymorphism and cancer susceptibility assessed by Begg's funnel plot and Egger's test (C vs. T). Log (OR): the natural logarithm of the odds ratio.
View Figure 2b
Heterogeneity analysis and publication bias risk
According to the results of heterogeneity analysis, no separate study shows influence on the pooled OR. Begg's funnel plot and Egger's test which were carried out to evaluate the publication bias risk. No publication bias was shown (Figure 2a, Figure 2b, Figure 2c and Figure 2d).
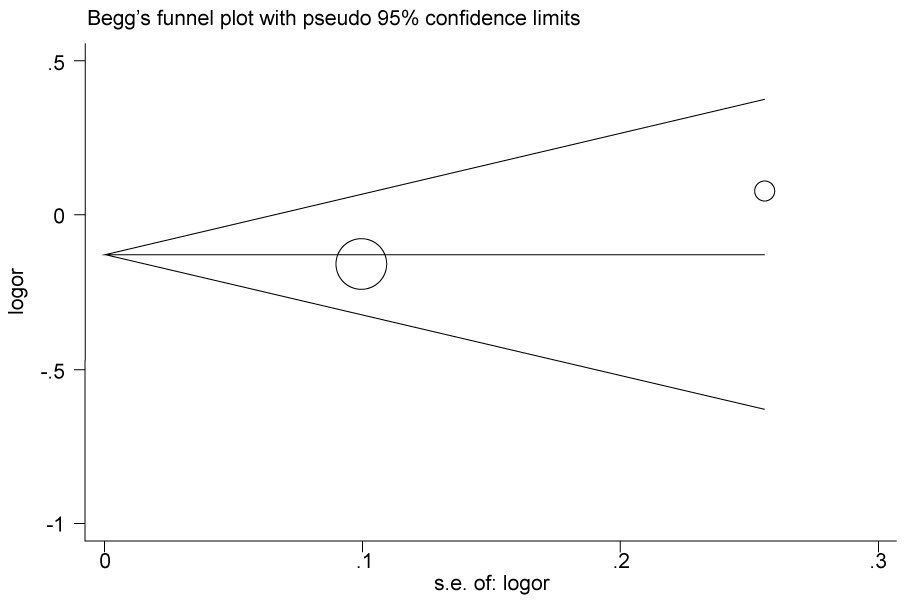
.
Figure 2c: Publication bias in studies of the association between the USF1 rs2774276 polymorphism and cancer susceptibility assessed by Begg's funnel plot and Egger's test (C vs. T). Log (OR): the natural logarithm of the odds ratio.
View Figure 2c
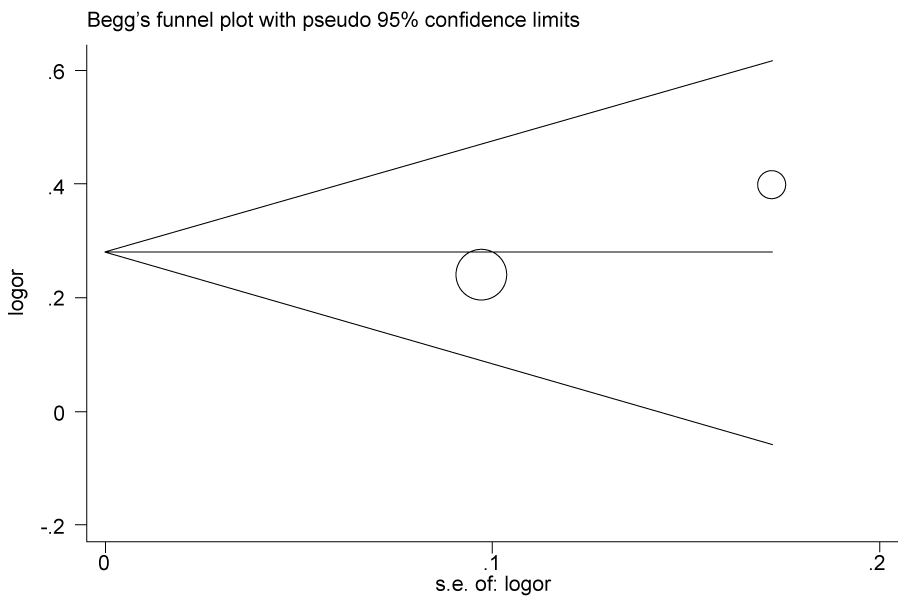
.
Figure 2d: Publication bias in studies of the association between the USF1 rs3737787 polymorphism and cancer susceptibility assessed by Begg's funnel plot and Egger's test (C vs. T). Log (OR): the natural logarithm of the odds ratio.
View Figure 2d
Discussion
A wide variety of genes are direct targets of USF, including genes involved in the immune response [28], glucid and lipid pathways [29], cell cycle [30,31], cell proliferation [32], and carcinogenesis [33-36]. Upstream transcription factors 1(USF1) is a member of the basic helix-loop-helix transcription factor family. It functions as a ubiquitous expressed transcription factor that regulates gene transcription by binding to the E-box motif of target genes [8,37]. According to plenty of studies, USF1 is one of the critical components within signal transduction path ways. Via regulating the expression of various genes they are involved in various aspects of cellular functions including regulation of cell growth and cell death. Therefore, disturbances within the proper function of USF1 may be related to tumorigenesis and cancer [38].
In a current study, Zhao et al. demonstrated that USF1 rs2516839 were an increased susceptibility of developing HCC in Chinese [22]. In addition, the other study conducted by Zhou et al. draw a similar conclusion. However, Zhou's study also demonstrated that USF1 rs3737787 is contributed to adding susceptibility of HCC [23]. While Yuan et al. proved that the USF1 alleles (rs2516838, rs3737787 and rs2516839) were all associated with HCC cancer susceptibility among Chinese [24]. Considering the inconsistent conclusions, we collect all the case-control studies currently published for a pooled analysis. In the present meta-analysis, we analyzed nine independent case-control studies with a sum of 1,927 cases and 4,037 controls. The pooled results illustrated that there is no evident association between the polymorphisms (rs2516838 and rs2774276) of USF1and cancer susceptibility. However, the polymorphisms (rs2516839 and rs3737787) were regarded as a risk factor separately. Moreover, in the stratified analysis, we found that the HCC cancer susceptibility is relevant with the polymorphism of USF1 rs2516839.
Although our present study have conducted a comprehensive retrieve for all the eligible studies, the pool results should be interpreted with caution due to several drawbacks in our meta-analysis. Firstly, the currently available case-control studies enrolled in our study were limited. Secondly, our study was conducted only in Chinese population, so the results may not be applied in other ethnicities. Thirdly, there was only one study in discussing the genetic predisposition of USF1 polymorphisms to PTC. We cannot evaluate any polymorphism of USF1on PTC susceptibility since eligible case-control studies that were currently published were insufficient for a pooled analysis. Finally, the effect of USF1polymorphisms on cancer susceptibility might be affected by some complex factors, such as age, gender, histological types of cancer, and matching criteria and so on. Therefore, more case-control studies are required to further assess gene association in cancer susceptibility.
To sum up, our work has revealed that USF1 rs2516839 and rs3737787polymorphisms alter the susceptibility of cancer. When considering a certain type of cancer, HCC appeared to be related to the USF12516839 polymorphism. A well-designed research may be needed to further explicit the relation between USF1 polymorphisms and cancer.
Declared of Interest
None
References
-
Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65: 5-29.
-
Chuang SC, La Vecchia C, Boffetta P (2009) Liver cancer: descriptive epidemiology and risk factors other than HBV and HCV infection. Cancer Lett 286: 9-14.
-
Ha NB, Ha NB, Ahmed A, Ayoub W, Daugherty TJ, et al. (2012) Risk factors for hepatocellular carcinoma in patients with chronic liver disease: a case-control study. Cancer Causes Control 23: 455-462.
-
Chen CJ, Yu MW, Liaw YF (1997) Epidemiological characteristics and risk factors of hepatocellular carcinoma. J Gastroenterol Hepatol 12: S294-308.
-
Kundu JK, Surh YJ (2008) Inflammation: gearing the journey to cancer. Mutat Res 659: 15-30.
-
Damiola F, Byrnes G, Moissonnier M, Pertesi M, Deltour I, et al. (2014) Contribution of ATM and FOXE1 (TTF2) to risk of papillary thyroid carcinoma in Belarusian children exposed to radiation. Int J Cancer 134: 1659-1668.
-
Bullock M, Duncan EL, O'Neill C, Tacon L, Sywak M, et al. (2012) Association of FOXE1 polyalanine repeat region with papillary thyroid cancer. J Clin Endocrinol Metab 97: E1814-1819.
-
Ikeda R, Nishizawa Y, Tajitsu Y, Minami K, Mataki H, et al. (2014) Regulation of major vault protein expression by upstream stimulating factor 1 in SW620 human colon cancer cells. Oncol Rep 31: 197-201.
-
Wu S, Mar-Heyming R, Dugum EZ, Kolaitis NA, Qi H, et al. (2010) Upstream transcription factor 1 influences plasma lipid and metabolic traits in mice. Hum Mol Genet 19: 597-608.
-
Putt W, Palmen J, Nicaud V, Tregouet DA, Tahri-Daizadeh N, et al. (2004) Variation in USF1 shows haplotype effects, gene : gene and gene : environment associations with glucose and lipid parameters in the European Atherosclerosis Research Study II. Hum Mol Genet 13: 1587-1597.
-
Galibert MD, Carreira S, Goding CR (2001) The Usf-1 transcription factor is a novel target for the stress-responsive p38 kinase and mediates UV-induced Tyrosinase expression. EMBO J 20: 5022-5031.
-
Corre S, Galibert MD (2005) Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res 18: 337-348.
-
Sirito M, Lin Q, Deng JM, Behringer RR, Sawadogo M (1998) Overlapping roles and asymmetrical cross-regulation of the USF proteins in mice. Proc Natl Acad Sci U S A 95: 3758-3763.
-
Jung HS, Kim KS, Chung YJ, Chung HK, Min YK, et al. (2007) USF inhibits cell proliferation through delay in G2/M phase in FRTL-5 cells. Endocr J 54: 275-285.
-
Muhlethaler-Mottet A, Di Berardino W, Otten LA, Mach B (1998) Activation of the MHC class II transactivator CIITA by interferon-gamma requires cooperative interaction between Stat1 and USF-1. Immunity 8: 157-166.
-
Pajukanta P, Lilja HE, Sinsheimer JS, Cantor RM, Lusis AJ, et al. (2004) Familial combined hyperlipidemia is associated with upstream transcription factor 1 (USF1). Nat Genet 36: 371-376.
-
Lee JC, Weissglas-Volkov D, Kyttala M, Sinsheimer JS, Jokiaho A, et al. (2007) USF1 contributes to high serum lipid levels in Dutch FCHL families and U.S. whites with coronary artery disease. Arterioscler Thromb Vasc Biol 27: 2222-2227.
-
Coon H, Xin Y, Hopkins PN, Cawthon RM, Hasstedt SJ, et al. (2005) Upstream stimulatory factor 1 associated with familial combined hyperlipidemia, LDL cholesterol, and triglycerides. Hum Genet 117: 444-451.
-
Holzapfel C, Baumert J, Grallert H, Müller AM, Thorand B, et al. (2008) Genetic variants in the USF1 gene are associated with low-density lipoprotein cholesterol levels and incident type 2 diabetes mellitus in women: results from the MONICA/KORA Augsburg case-cohort study, 1984-2002. Eur J Endocrinol 159: 407-416.
-
Laurila PP, Naukkarinen J, Kristiansson K, Ripatti S, Kauttu T, et al. (2010) Genetic association and interaction analysis of USF1 and APOA5 on lipid levels and atherosclerosis. Arterioscler Thromb Vasc Biol 30: 346-352.
-
Kristiansson K, Ilveskoski E, Lehtimäki T, Peltonen L, Perola M, et al. (2008) Association analysis of allelic variants of USF1 in coronary atherosclerosis. Arterioscler Thromb Vasc Biol 28: 983-989.
-
Zhao X, Wang T, Liu B, Wu Z, et al. (2015) Yu S Significant association between upstream transcription factor 1 rs2516839 polymorphism and hepatocellular carcinoma risk: a case-control study. Tumour Biol 36: 2551-2558.
-
Zhou X, Zhu HQ, Ma CQ, Li HG, Liu FF, et al. (2014) Two polymorphisms of USF1 gene (-202G>A and -844C>T) may be associated with hepatocellular carcinoma susceptibility based on a case-control study in Chinese Han population. Med Oncol 31: 301.
-
Yuan Q, Bu Q, Li G, Zhang J, Cui T, et al. (2015) Association between single nucleotide polymorphisms of upstream transcription factor 1 (USF1) and susceptibility to papillary thyroid cancer. Clin Endocrinol (Oxf).
-
Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327: 557-560.
-
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719-748.
-
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177-188.
-
Galibert MD, Boucontet L, Goding CR, Meo T (1997) Recognition of the E-C4 element from the C4 complement gene promoter by the upstream stimulatory factor-1 transcription factor. J Immunol 159: 6176-6183.
-
Roth U, Jungermann K, Kietzmann T (2004) Modulation of glucokinase expression by hypoxia-inducible factor 1 and upstream stimulatory factor 2 in primary rat hepatocytes. Biol Chem 385: 239-247.
-
North S, Espanel X, Bantignies F, Viollet B, Vallet V, et al. (1999) Regulation of cdc2 gene expression by the upstream stimulatory factors (USFs). Oncogene 18: 1945-1955.
-
Pawar SA, Szentirmay MN, Hermeking H, Sawadogo M (2004) Evidence for a cancer-specific switch at the CDK4 promoter with loss of control by both USF and c-Myc. Oncogene 23: 6125-6135.
-
Goueli BS, Janknecht R (2003) Regulation of telomerase reverse transcriptase gene activity by upstream stimulatory factor. Oncogene 22: 8042-8047.
-
Hale TK, Braithwaite AW (1995) Identification of an upstream region of the mouse p53 promoter critical for transcriptional expression. Nucleic Acids Res 23: 663-669.
-
Reisman D, Rotter V (1993) The helix-loop-helix containing transcription factor USF binds to and transactivates the promoter of the p53 tumor suppressor gene. Nucleic Acids Res 21: 345-350.
-
Davis PL, Miron A, Andersen LM, Iglehart JD, Marks JR (1999) Isolation and initial characterization of the BRCA2 promoter. Oncogene 18: 6000-6012.
-
Jaiswal AS, Narayan S (2001) Upstream stimulating factor-1 (USF1) and USF2 bind to and activate the promoter of the adenomatous polyposis coli (APC) tumor suppressor gene. J Cell Biochem 81: 262-277.
-
Chen B, Hsu R, Li Z, Kogut PC, Du Q, et al. (2012) Upstream stimulatory factor 1 activates GATA5 expression through an E-box motif. Biochem J 446: 89-98.
-
Horbach T, Götz C2, Kietzmann T3, Dimova EY3 (2015) Protein kinases as switches for the function of upstream stimulatory factors: implications for tissue injury and cancer. Front Pharmacol 6: 3.





